Proteomics Data Analysis (PDA)
Course Description
Mass spectrometry based proteomic experiments generate ever larger datasets and, as a consequence, complex data interpretation challenges. This course focuses on bioinformatics and statistics for peptide identification, quantification, and differential analysis. Moreover, more advanced experimental designs and blocking will also be introduced. The core focus will be on shotgun proteomics data, and quantification using label-free precursor peptide (MS1) ion intensities. The course will rely exclusively on free and user-friendly software, all of which can be directly applied in your lab upon returning from the course. You will also learn how to submit data to PRIDE/ProteomeXchange, which is a common requirement for publication in the field, and how to browse and reprocess publicly available data from online repositories. The course will provide a solid basis for beginners, but will also bring new perspectives to those already familiar with standard data interpretation procedures in proteomics.
Target Audience
This course is oriented towards biologists and bioinformaticians with a particular interest in differential analysis for quantitative proteomics.
Prerequisites
The participants should have a basic knowledge about mass spectrometry based proteomics. Experience in analysing proteomics data is an advantage, but not mandatory. The course does not require advanced computer skills.
You will also be requested to watch the following videos before attending the course: Mass Spectrometry and Bioinformatics for Proteomics
Detailed Program
1. Bioinformatics for Proteomics
2. Statistics for Proteomics Data Analysis
2.0. Software and Data
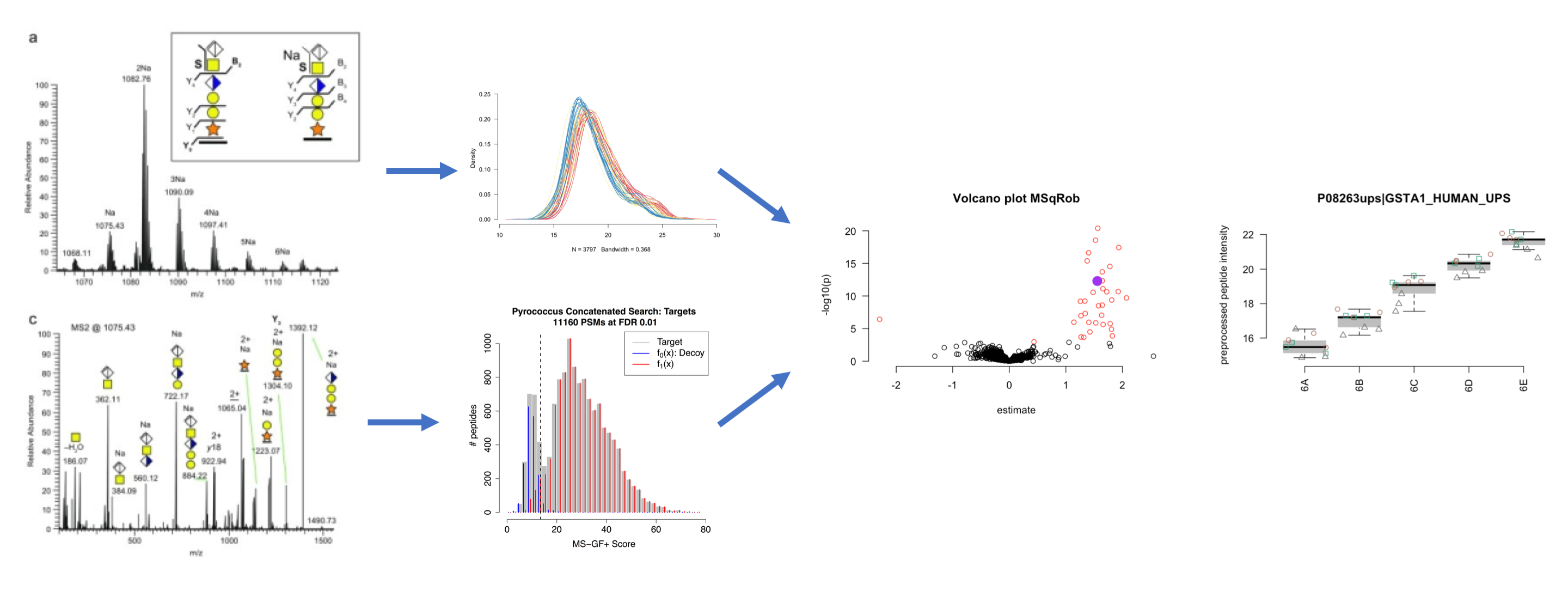
More information on our tools can be found in our papers (L. J. Goeminne, Gevaert, and Clement 2016), (L. J. E. Goeminne et al. 2020) and (Sticker et al. 2020). Please refer to our work when using our tools.
2.1. Identification
- Target Decoy QC
- Tutorial
- Solutions: Uniprot search, Swissprot search
2.2. Statistical Data Analysis of Label Free Quantitative
- Preprocessing & Analysis of Label Free Quantitative Proteomics Experiments with Simple Designs
- Lecture: html, pdf
- Tutorial: preprocessing
- Wrap-up Peptide-based Models: html, pdf
- Statistical Inference & Analysis of Experiments with Factorial Designs
Instructors
License
This project is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)
