Introduction to TargetDecoy
Lieven Clement
Ghent Universitylibrary(TargetDecoy)
library(ggplot2)1 Introduction
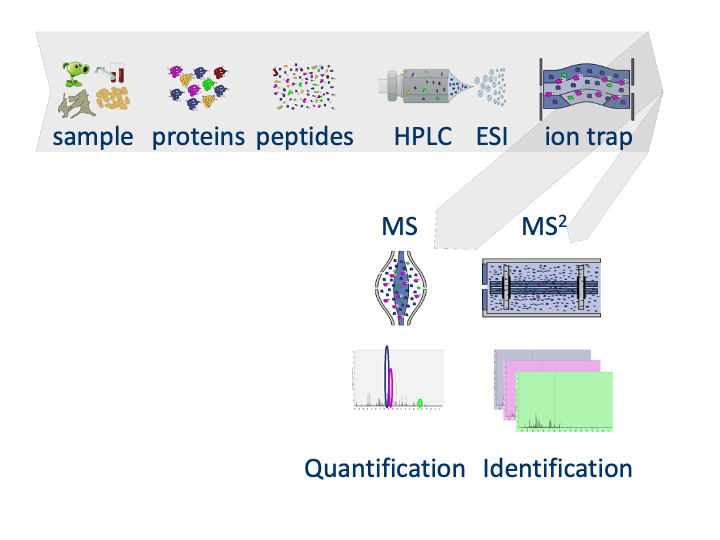
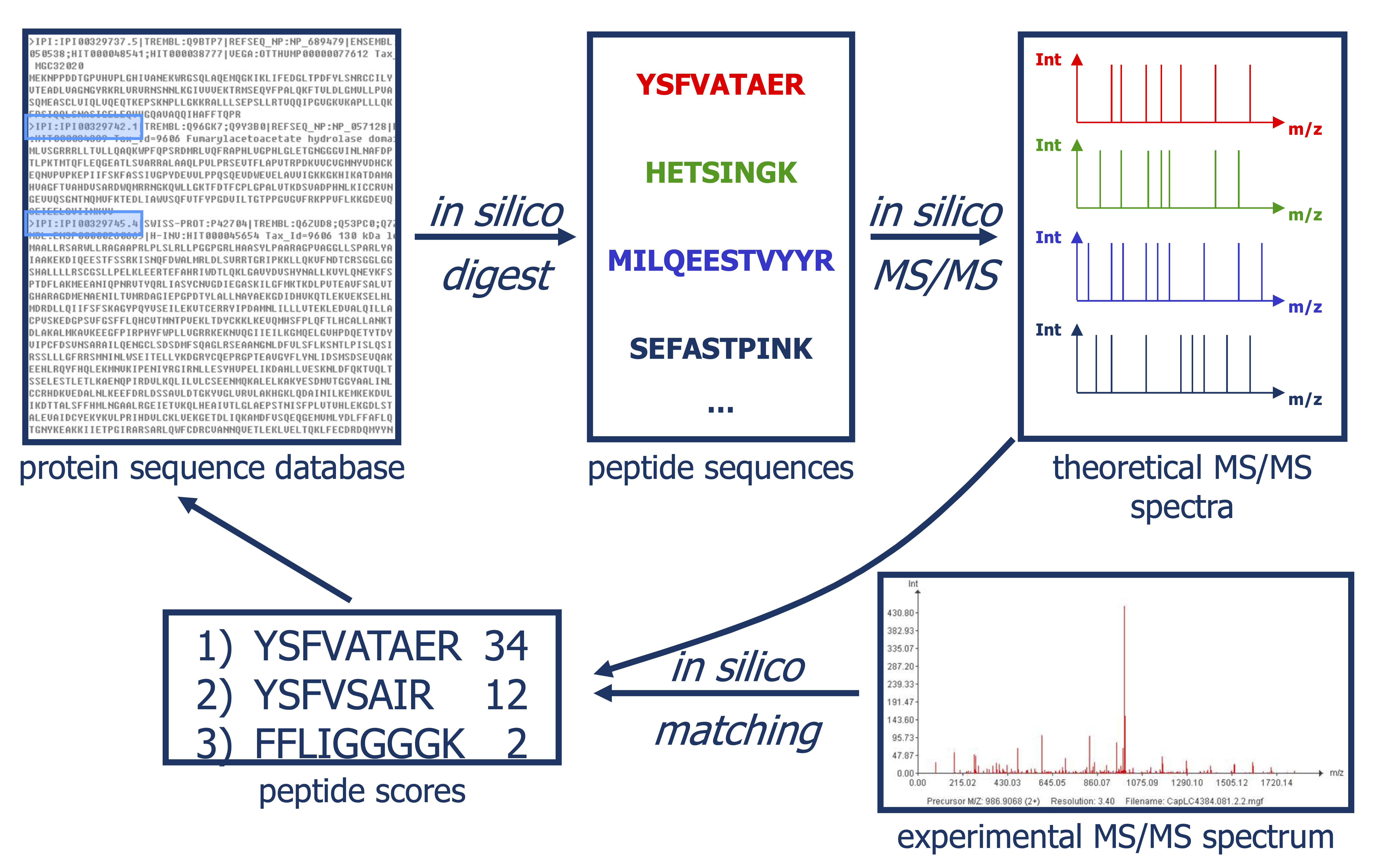 Slide courtesy to Lennart Martens
Slide courtesy to Lennart Martens
1.1 Why did Lennart mention that “statistics were out of the window”
library(TargetDecoy)
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──
#> ✔ tibble 3.1.7 ✔ dplyr 1.0.9
#> ✔ tidyr 1.2.0 ✔ stringr 1.4.0
#> ✔ readr 2.1.2 ✔ forcats 0.5.1
#> ✔ purrr 0.3.4
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
data("ModSwissXT")
hlp <- TargetDecoy:::.getDF(ModSwissXT)
names(hlp) <- names(hlp) %>%
str_replace(pattern=":",replacement="_")
hlp <- hlp %>%
mutate(omssa_evalue=as.double(omssa_evalue))E-values: Probability that a random candidate peptide produces a higher score than the observed PSM score.
Probability that a random candidate peptide produces a higher score that the observed PSM score for a real search with OMSSA.
library(TargetDecoy)
library(tidyverse)
data("ModSwissXT")
hlp <- TargetDecoy:::.getDF(ModSwissXT)
names(hlp) <- names(hlp) %>%
str_replace(pattern=":",replacement="_")
hlp <- hlp %>%
mutate(omssa_evalue=as.double(omssa_evalue))
hlp %>%
filter(!is.na(omssa_evalue)) %>%
ggplot(aes(omssa_evalue)) +
geom_histogram(breaks=seq(0,100,5)) +
xlab("E-value (%)") +
ylab("frequency")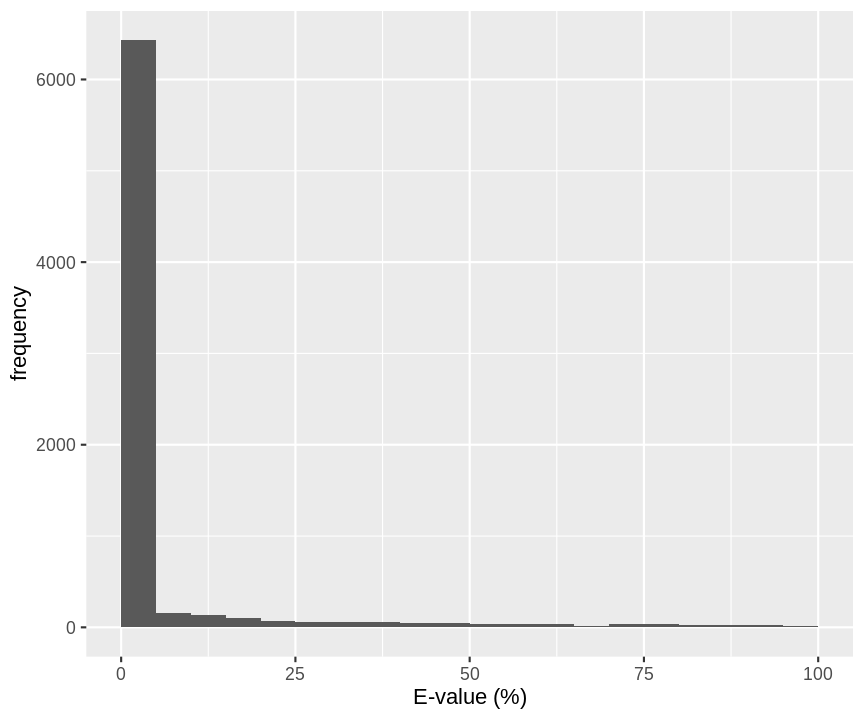
- E-value we expect for an observed spectrum and a random candidate peptide.
data.frame(evalue=runif(20000,0,100)) %>%
ggplot(aes(evalue)) +
geom_histogram(breaks=seq(0,100,5)) +
xlab("E-value (%)") +
ylab("frequency")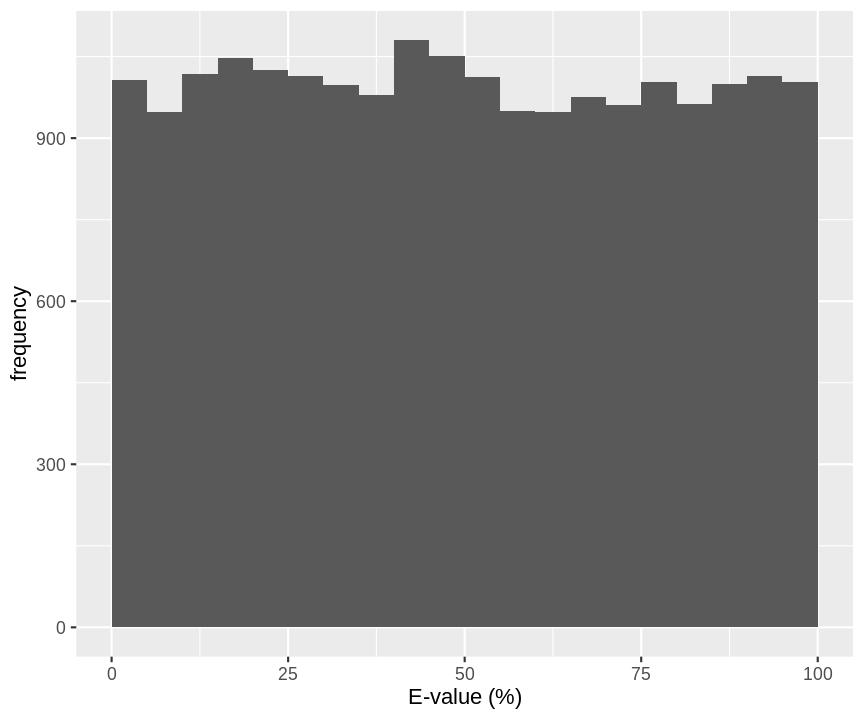
- Probability that a random candidate peptide produces a higher score than the observed PSM score for decoys in a real search with OMSSA.
hlp %>%
filter(isdecoy & !is.na(omssa_evalue)) %>%
ggplot(aes(omssa_evalue)) +
geom_histogram(breaks=seq(0,100,5)) +
xlab("E-value (%)") +
ylab("frequency")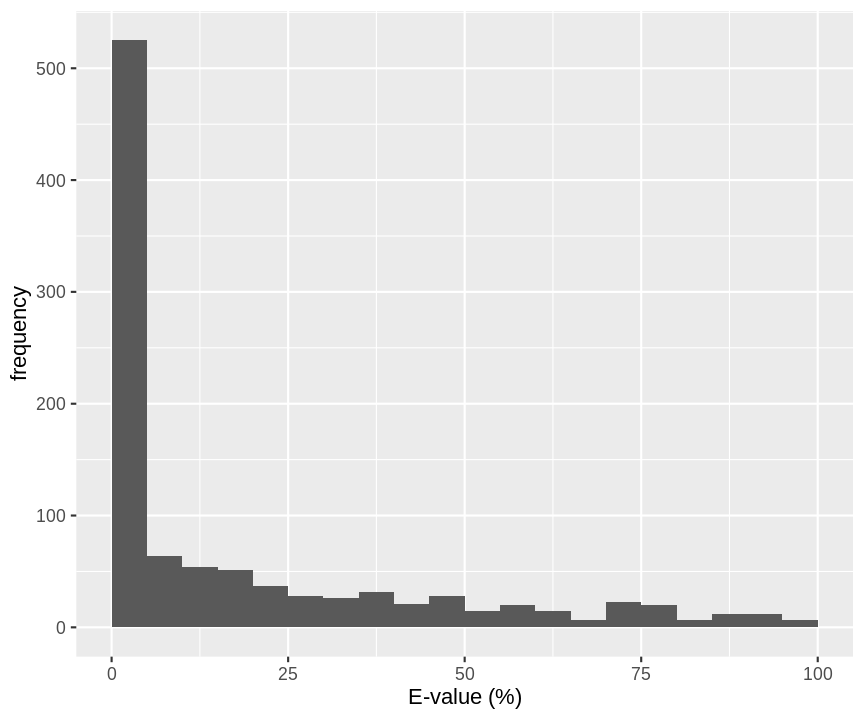
A bad hit is the random hit with the best score so it is also bound to have a low E-value.
If we look at E-values for all PSMs they are only useful as a score.
We should know the distribution of the maximum score of random candidate peptides when we want to do the statistics.
hlp %>%
filter(!is.na(omssa_evalue)) %>%
ggplot(aes(-log10(omssa_evalue))) +
geom_histogram(breaks=seq(0,1000,.5)) +
xlab("-log10(E-value)") +
ylab("frequency") +
xlim(0,20) 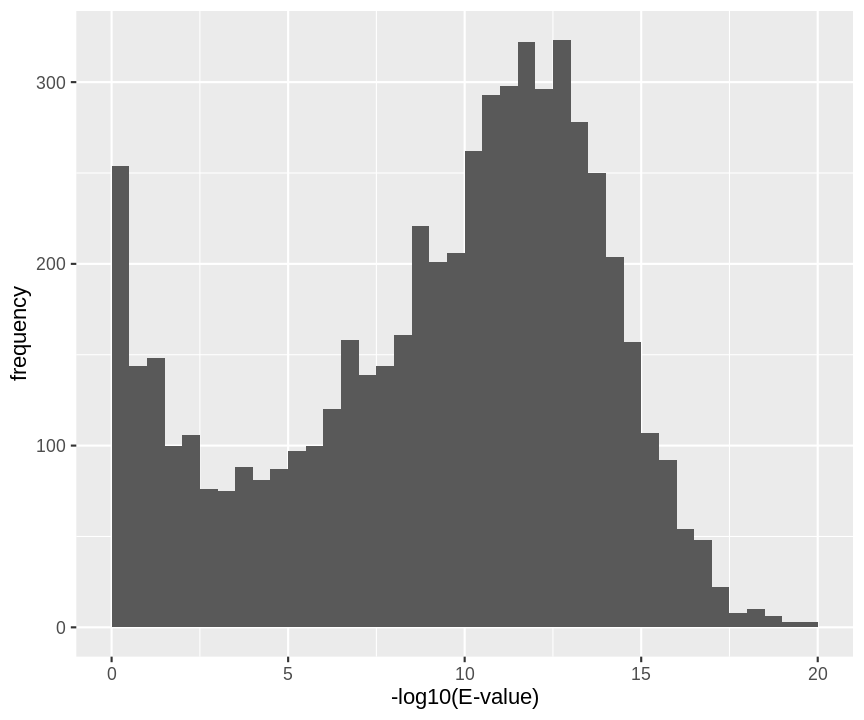
2 Concepts
2.1 Basic Statistical Concepts
names(hlp) <- names(hlp) %>% str_replace(pattern="-",replacement = "_")
hlp <- hlp %>% mutate(ms_gf_specevalue=as.double(ms_gf_specevalue))library(mgcv)
#> Loading required package: nlme
#>
#> Attaching package: 'nlme'
#> The following object is masked from 'package:dplyr':
#>
#> collapse
#> This is mgcv 1.8-40. For overview type 'help("mgcv-package")'.
dec <- -log10(hlp$ms_gf_specevalue[hlp$isdecoy]) %>% na.exclude()
tar <- -log10(hlp$ms_gf_specevalue[!hlp$isdecoy]) %>% na.exclude()
breaks <- seq(0,30,.5)
#binWidth <-2
#breaks <- seq(floor(min(c(dec,tar))/binWidth)*binWidth,ceiling(max(c(dec,tar))/binWidth)*binWidth,binWidth)
#code if we register the modes by substracting the mode from the target scores and the decoy scores.
#breaks=seq(-(ceiling(abs(min(c(dec,tar))/binWidth))+.5)*binWidth,(ceiling(max(c(dec,tar))/binWidth)+.5)*binWidth,binWidth)
histDec <- hist(dec,breaks=breaks,plot = FALSE)
histTar <- hist(tar,breaks=breaks,plot=FALSE)
histSam <- hist(c(dec,tar),breaks=breaks, plot = FALSE)
grid<-seq(0,30,.1)
countsTarG<-data.frame(y=histTar$counts-histDec$counts,x=histTar$mids)
countsTarG$y[countsTarG$y<0]<-0
fitTarG<-gam(y~s(x),data=countsTarG,family=poisson)
fitTarGrid<-exp(predict(fitTarG,newdata=data.frame(x=grid)))
countsDec<-data.frame(y=histDec$counts,x=histDec$mids)
fitDec<-gam(y~s(x),data=countsDec,family=poisson)
fitSamBad<-exp(predict(fitDec,newdata=data.frame(x=grid)))*2
plot(histSam,xlab="MS-GF+ Score",ylab="# PSMs",main="Pyrococcus Search",border="white",col="grey",cex.axis=1.5,cex.main=1.5,cex.lab=1.5,ylim=c(0,1500), axes =FALSE)
axis(side=2,at=c(0,750,1500))
axis(side=1,at=c(0,10,20,30))
lines(grid,fitSamBad+fitTarGrid,col="black",lwd=2)
lines(grid,fitSamBad,col="#FF9900",lwd=2)
lines(grid,fitTarGrid,col="#009900",lwd=2)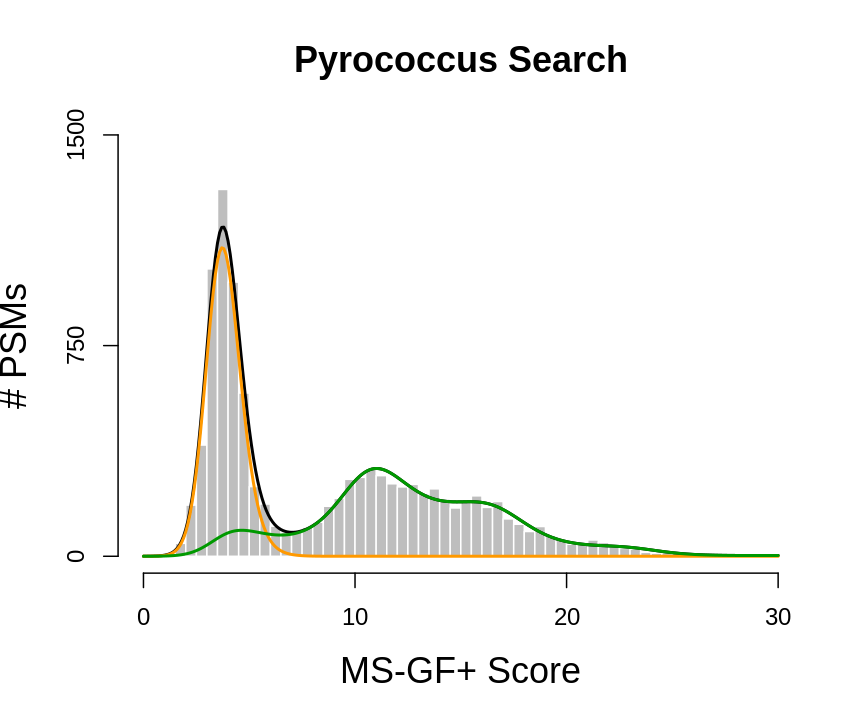
Let \(x\) be the PSM score
The scores will follow a mixture distribution:
\[f(x) = \pi_b \mathbin{\color{orange}{ f_b(x)}}+(1-\pi_b)\mathbin{\color{green}{ f_g(x)}},\] The local fdr is also referred to as the posterior error probability (PEP), and is the probability that a PSM with a score \(x\) is a bad hit.
\[ \begin{array}{lcl} \text{lfdr}(x) &=&\text{PEP(x)}\\ &=& \text{P}[\text{bad hit} \vert X=x]\\ &=& \frac{\pi_b f_b(x)}{f(x)} \end{array} \]
We will return a list of PSMs by using the FDR:
\[FDR = E\left[\frac{FP}{TP + FP}\right]\]
- FP: number of false positives, bad hits
- TP: number of true positives, good hits
\[ \begin{array}{lcl} \text{FDR}(x\geq t) &=& \text{P}[\text{bad hit }\vert X \geq t]\\\\ &=&\frac{\pi_b\int\limits_{x=t}^{+\infty}f_b(x)}{\int\limits_{x=t}^{+\infty}f(x)}\\\\ &=&\frac{\pi_b[1-F_b(t)]}{1-F(t)} \end{array} \]
- So the FDR is a set property, it measure the probability on a bad hit in the set of PSMs with scores \(X\geq t\).
Our list, thus consists of all PSMs with a score \(x\) above a threshold t.
plot(histSam,xlab="MS-GF+ Score",ylab="# PSMs",main="Pyrococcus Search",border="white",col="grey",cex.axis=1.5,cex.main=1.5,cex.lab=1.5,ylim=c(0,1500), axes =FALSE)
axis(side=2,at=c(0,750,1500))
axis(side=1,at=c(0,10,20,30))
lines(grid,fitSamBad+fitTarGrid,col="black",lwd=2)
lines(grid,fitSamBad,col="#FF9900",lwd=2)
lines(grid,fitTarGrid,col="#009900",lwd=2)
text(pos=4,5,1430,label=expression(x >= t),col="darkorchid4",cex=2)
rect(5,-10,30,1500,lwd=2,border="darkorchid4")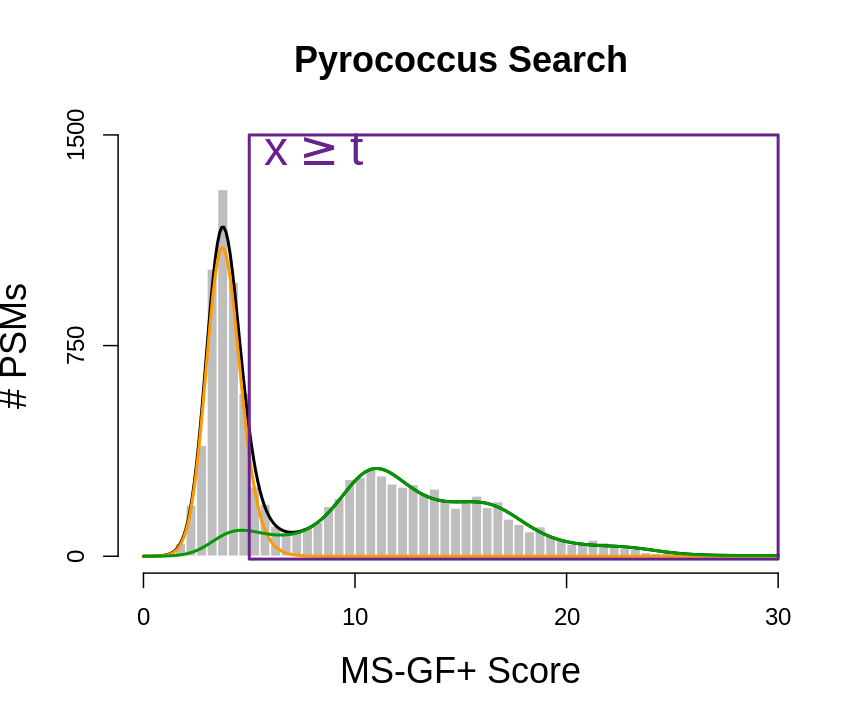
- So we know how many PSMs we return, i.e. TP + FP: \[\text{#PSMs with } x \geq t\]
and we can also estimate the probability on a PSM above the threshold empirically:
\[1-\hat{\text{F}}(t) = \frac{\text{#PSMs with } x \geq t}{\text{#PSMs}}\]
- So to estimate the FDR we only have to estimate the expected number of PSMs that are bad hits with a score \(x\) above the threshold \(t\).
\[\widehat{\text{FDR}}(t) = \frac{E\left[\#\text{Bad PSM hits with } X \geq t\right]}{\text{#PSMs with } x \geq t}\]
2.2 Competitive target decoy approach
- Search against decoy database to generate representative bad hits
- Reverse database is popular
- Concatenated search is most popular
- Advantage, a number of bad hits already matches with decoys
\(\rightarrow\) we know that these are bad hits
\(\rightarrow\) we have to infer on less target PSMs.
hlp %>%
filter(!is.na(ms_gf_specevalue)) %>%
ggplot(aes(-log10(ms_gf_specevalue))) +
geom_histogram(breaks=seq(0,1000,.5)) +
xlab("-log10(E-value)") +
ylab("frequency") +
xlim(0,40) +
facet_grid(isdecoy~.)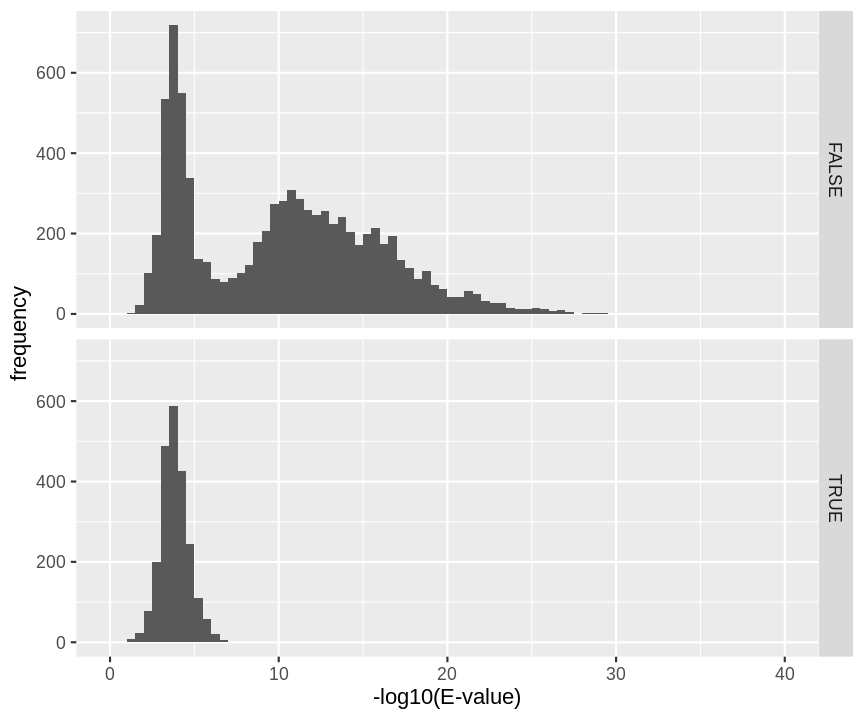
We estimate that by using the decoys: \[\text{# Decoys with x} \geq t\] So our estimated FDR becomes
\[\widehat{\text{FDR}}(x\geq t) = \frac{\text{# Decoys} \geq t}{\text{# Targets} \geq t}\] If we rewrite the FDR we can see the TDA assumptions:
\[ \begin{array}{lcl} \widehat{\text{FDR}}(x\geq t)&=&\frac{\text{# Decoys} \geq t}{\text{# Targets} \geq t}\\\\ &=&\frac{\frac{\text{# Decoys}}{\text{# Targets}}\frac{\text{# Decoys with }x \geq t}{\text{# Decoys}}}{\frac{\text{# Targets with } x \geq t}{\text{# Targets}}}\\\\ &=&\frac{\frac{\hat{\text{E}}\left[\text{# Bad Targets}\right]}{\text{# Targets}}\frac{\hat{\text{E}}\left[\text{# Bad Targets with }X \geq t\right]}{\hat{\text{E}}\left[\text{# Bad targets}\right]}}{\frac{\text{# Targets with } x \geq t}{\text{# Targets}}} \\\\ &=&\frac{\hat{\text{P}}\left[\text{Bad Target} \right]\times\hat{\text{P}}\left[\text{Bad Target}\vert X \geq t \right]}{\hat{\text{P}}\left[\text{Target}\vert X \geq t\right]}\\\\ &=&\frac{\hat\pi_b [1-\hat{F}_b(t)]}{1-\hat{F}(t)} \end{array} \]
So the TDA has the following assumptions:
A bad hit is equally likely to match to a decoy as to a target sequence. \(\rightarrow\) we can thus estimate the fraction of bad hits or the probability on a bad hit as \[\hat{\pi}_b = \frac{\# \text{Decoys}}{\# \text{Targets}}\]
Bad target PSM scores and decoy PSM scores are equaly distributed.
3 Diagnostic plots for the TDA
We will evaluate the TDA assumptions using diagnostic plots that compares the empirical distribution of decoy and target PSM scores.
- Histograms
- P-P plots
With P-P plots will plot for each observed PSM score \(t\) the empirical probability to observe a decoy with score \(x \leq t\) to the empirical probability to observe a target with score \(x \leq t\):
\[\hat{\text{P}}\left[\text{decoy with score } x \leq t\right] = \frac{\# \text{decoys with score } x \leq t}{\# \text{decoys}}\]
\[ \hat{\text{P}}\left[\text{target with score } x \leq t\right] = \frac{\# \text{target with score } x \leq t}{\# \text{targets}} \]
If the two distributions are the same the dots of the P-P plot should follow the 1-1 line.
This will not be the case. We expect the distribution of the target PSMs:
- to be similarly distributed as the decoys for low scores
- and enriched with many high scores corresponding to target PSMs which are matching to the proper peptide sequence in the data base.
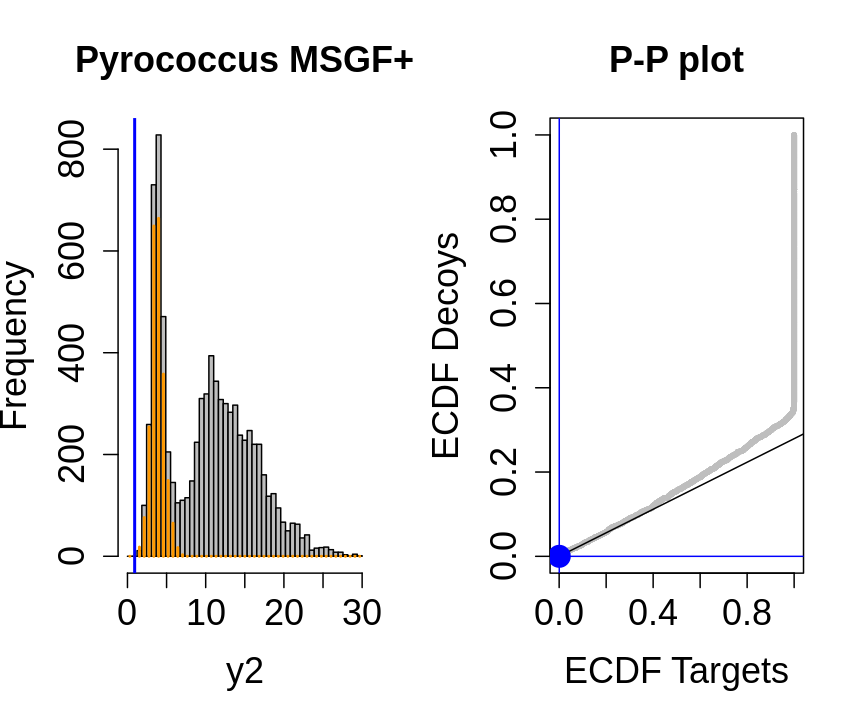
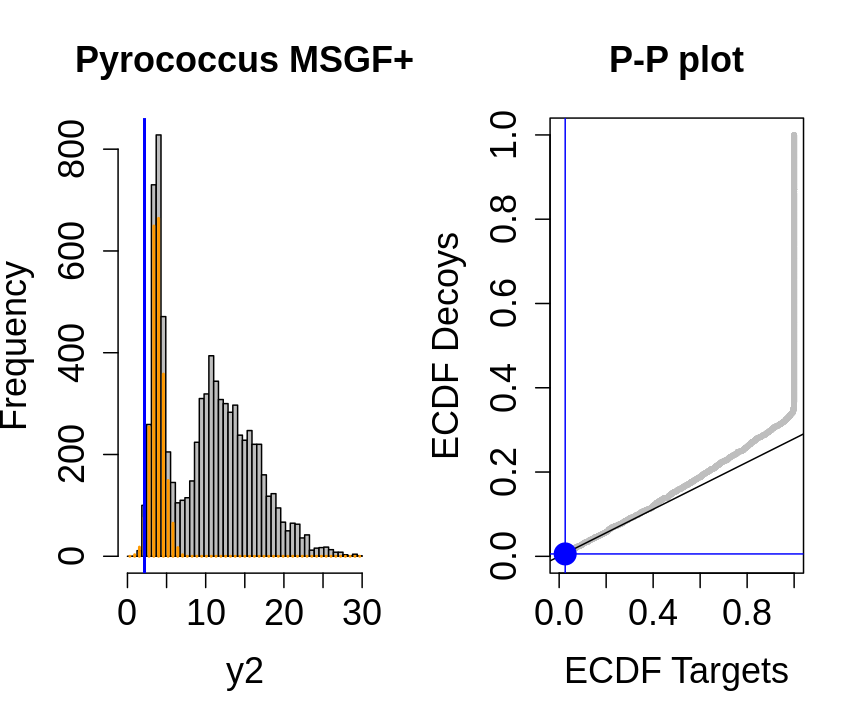
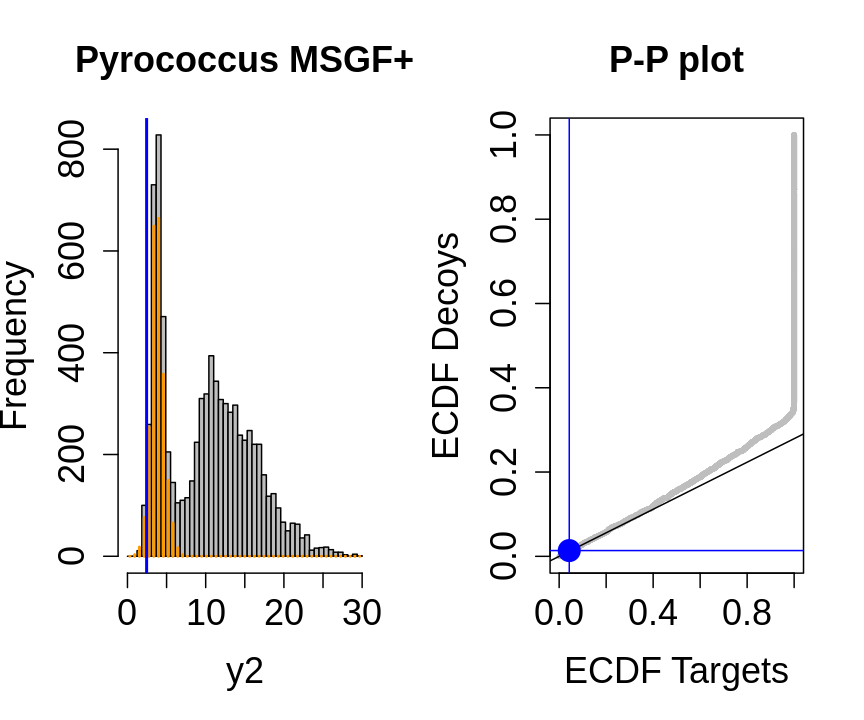
hlp <- hlp %>%
mutate(score=-log10(hlp$ms_gf_specevalue))
y1<-hlp$score[hlp$isdecoy] %>% na.exclude
y2<-hlp$score[!hlp$isdecoy] %>% na.exclude
F1<-ecdf(y1)
F2<-ecdf(y2)
breaks<-seq(floor(min(c(y1,y2))),ceiling(max(c(y1,y2))),length.out=50)
pi0<-length(y1)/length(y2)
for (x in quantile(c(y1,y2),c(0,.01,.02,seq(0.1,1,.1))))
{
par(mfrow=c(1,2))
hist(y2,breaks=breaks,main="Pyrococcus MSGF+",cex.axis=1.5,cex.lab=1.5,cex.main=1.5,col="grey")
decHist<-hist(y1,breaks=breaks,plot=FALSE)
points(decHist$mids,decHist$counts,col="#FF9900",type="h",lwd=2)
abline(v=x,col="blue",lwd=2)
plot(F1(y2),F2(y2),xlab="ECDF Targets",ylab="ECDF Decoys",cex=.4,main="P-P plot",cex.axis=1.5,cex.lab=1.5,cex.main=1.5,col="grey",pch=19)
abline(a=0,b=pi0)
abline(v=F1(x),col="blue")
abline(h=F2(x),col="blue")
points(F1(x),F2(x),col="blue",cex=2,pch=19)
}
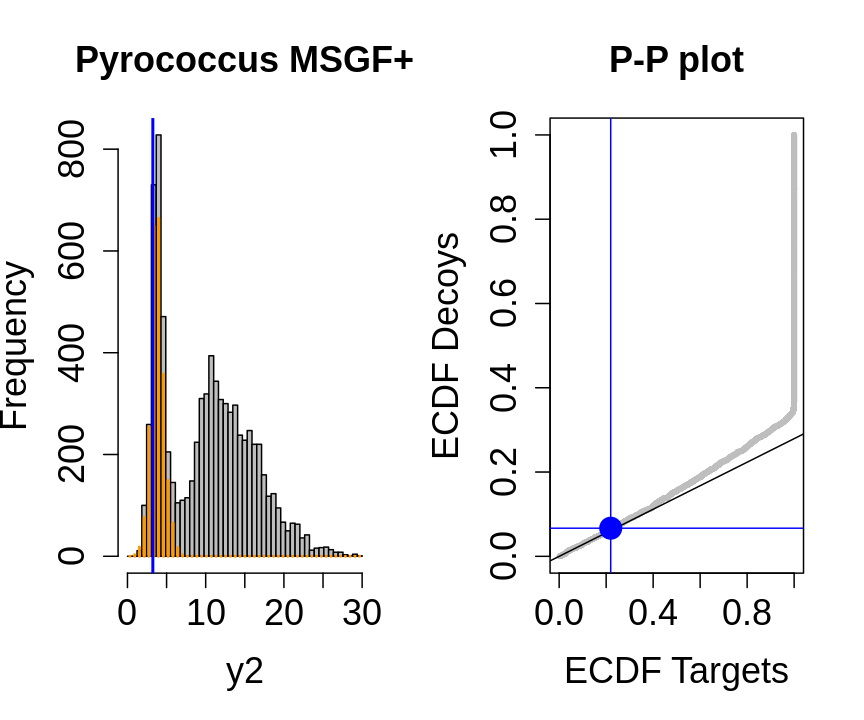
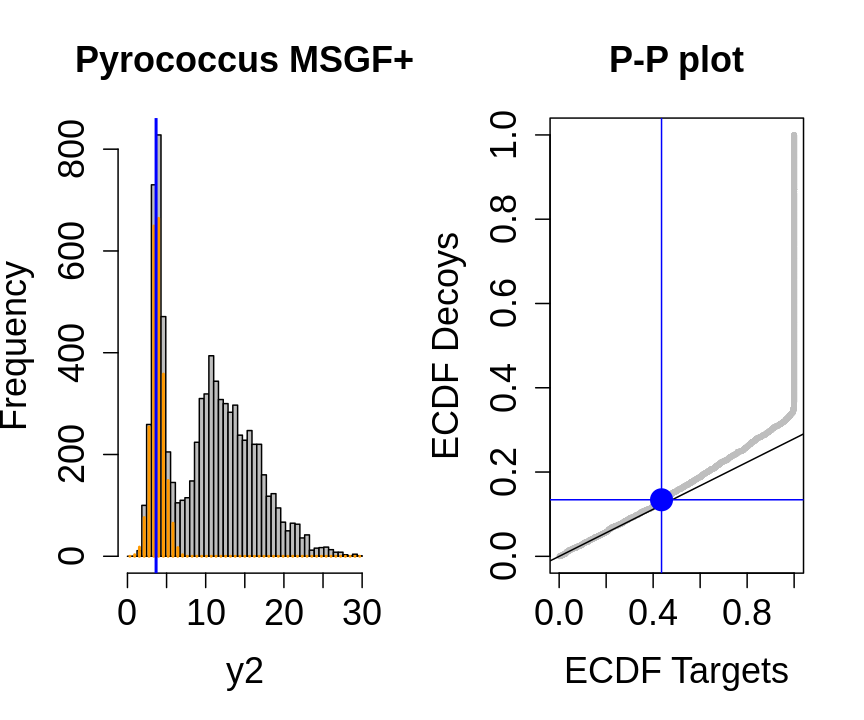
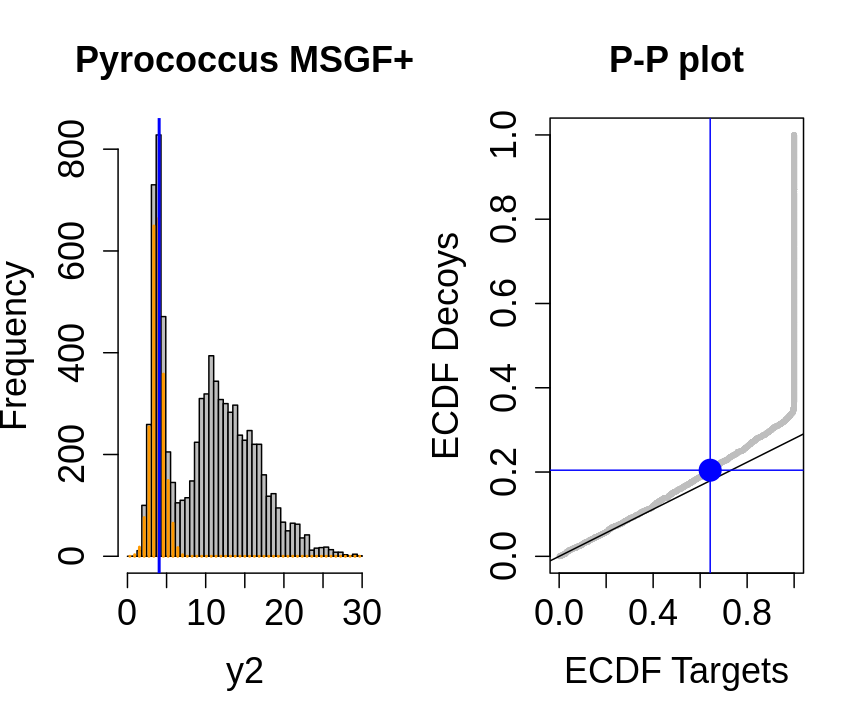
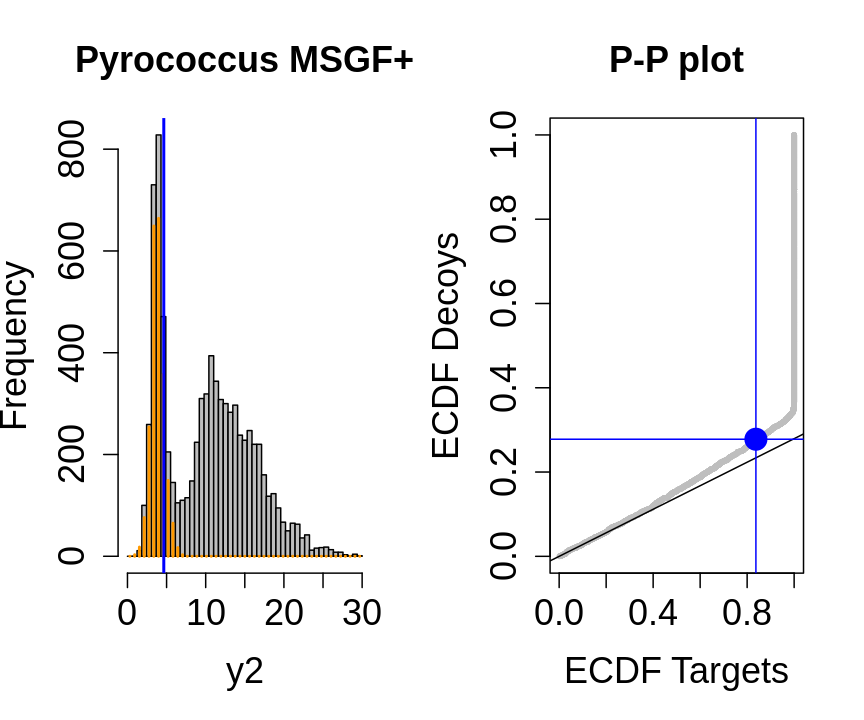
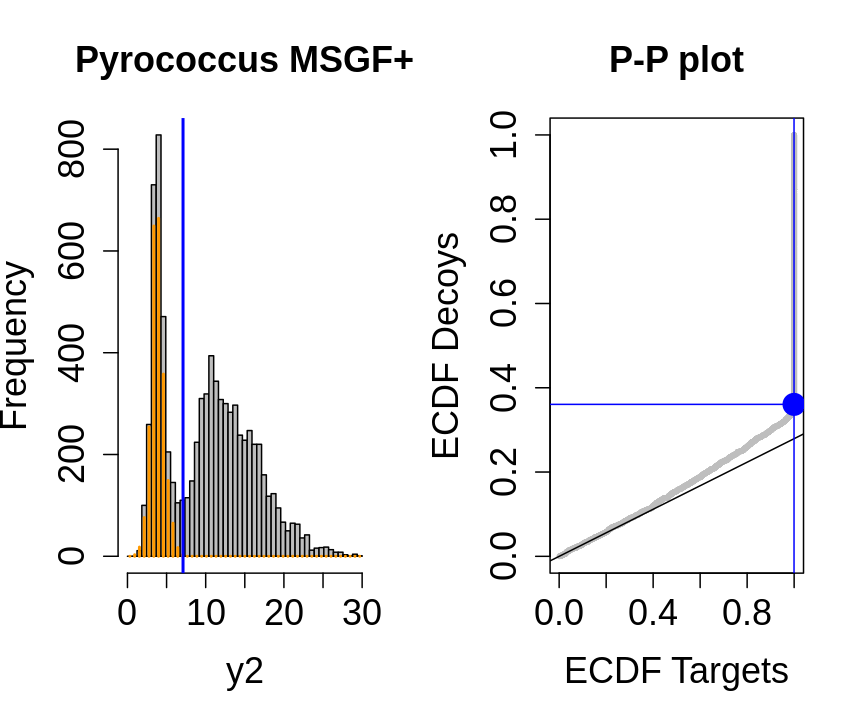
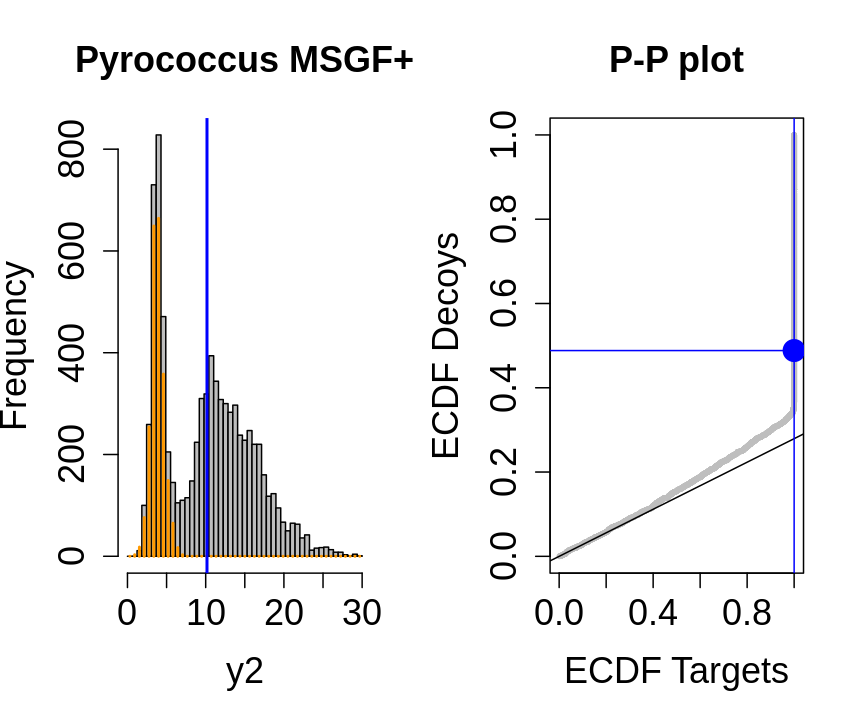
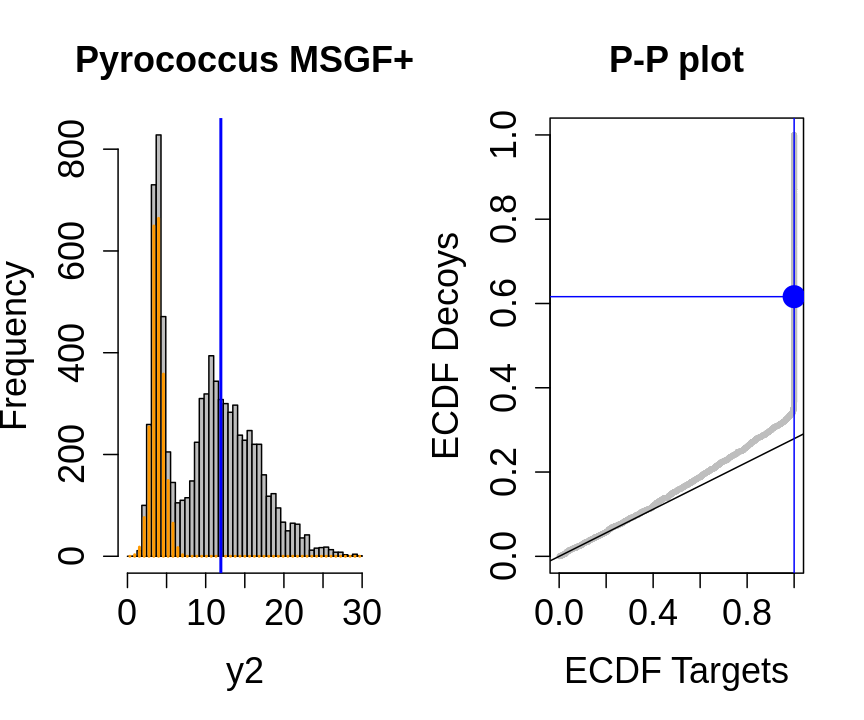
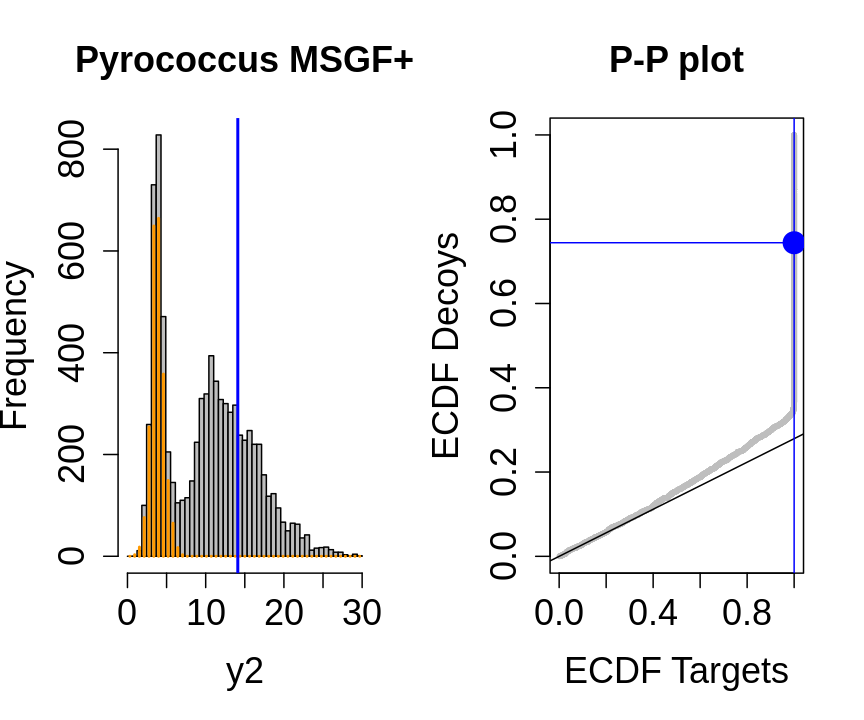
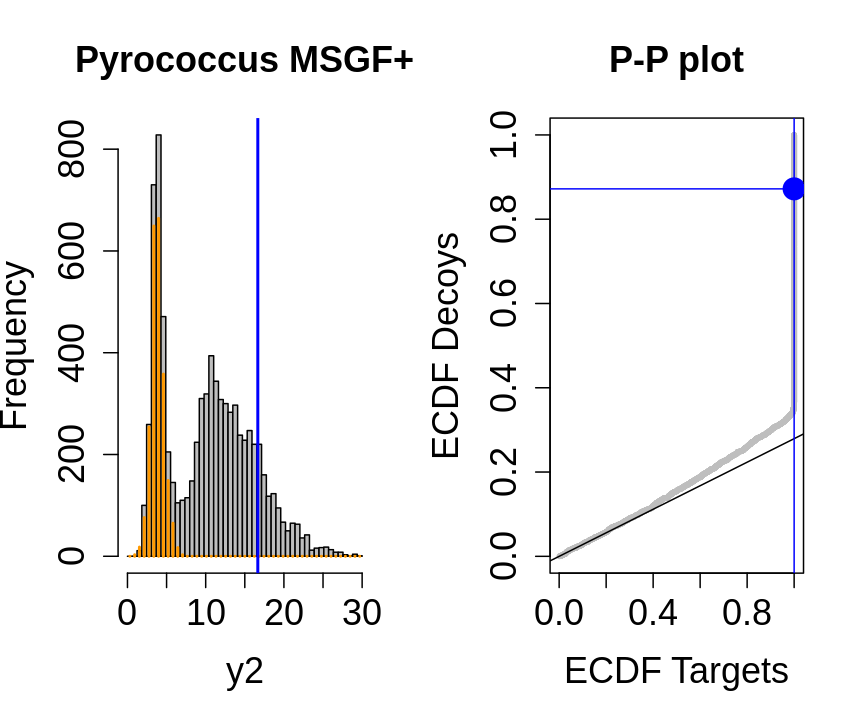
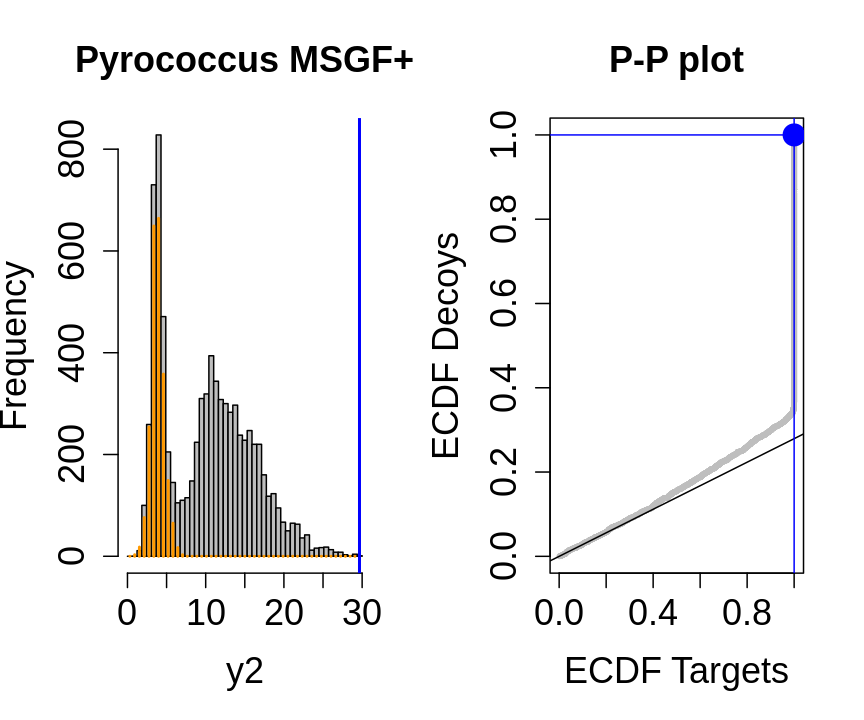
Note, that
- the points corresponding to low score values follow a straight line indicating that targets and decoys scores are similarly distributed
- this line has an angle equal to fraction of bad hits, which is estimated as \(\hat\pi_b=\frac{\# \text{decoys}}{\#\text{targets}}\) and is indicated by the black line in the plot.
- This line can be used to assess the assumption that bad hits are equally likely matching to targets sequences as to decoy sequences.