Example: MSGF+ search on Swiss-Prot
Lieven Clement
Ghent University1 Load Libraries
## Warning in fun(libname, pkgname): mzR has been built against a different Rcpp version (1.0.12)
## than is installed on your system (1.0.13.1). This might lead to errors
## when loading mzR. If you encounter such issues, please send a report,
## including the output of sessionInfo() to the Bioc support forum at
## https://support.bioconductor.org/. For details see also
## https://github.com/sneumann/mzR/wiki/mzR-Rcpp-compiler-linker-issue.2 Download data in working directory
3 Load Data in R
## reading pyrococcusMSGF+.mzid...## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE
## Warning in type.convert.default(...): 'as.is' should be specified by the
## caller; using TRUE## DONE!4 Launch the Shiny Gadget
Explore the results for search eninge scores to find correct names of search engine scores in the mzID.
5 Evaluate target decoy assumptions
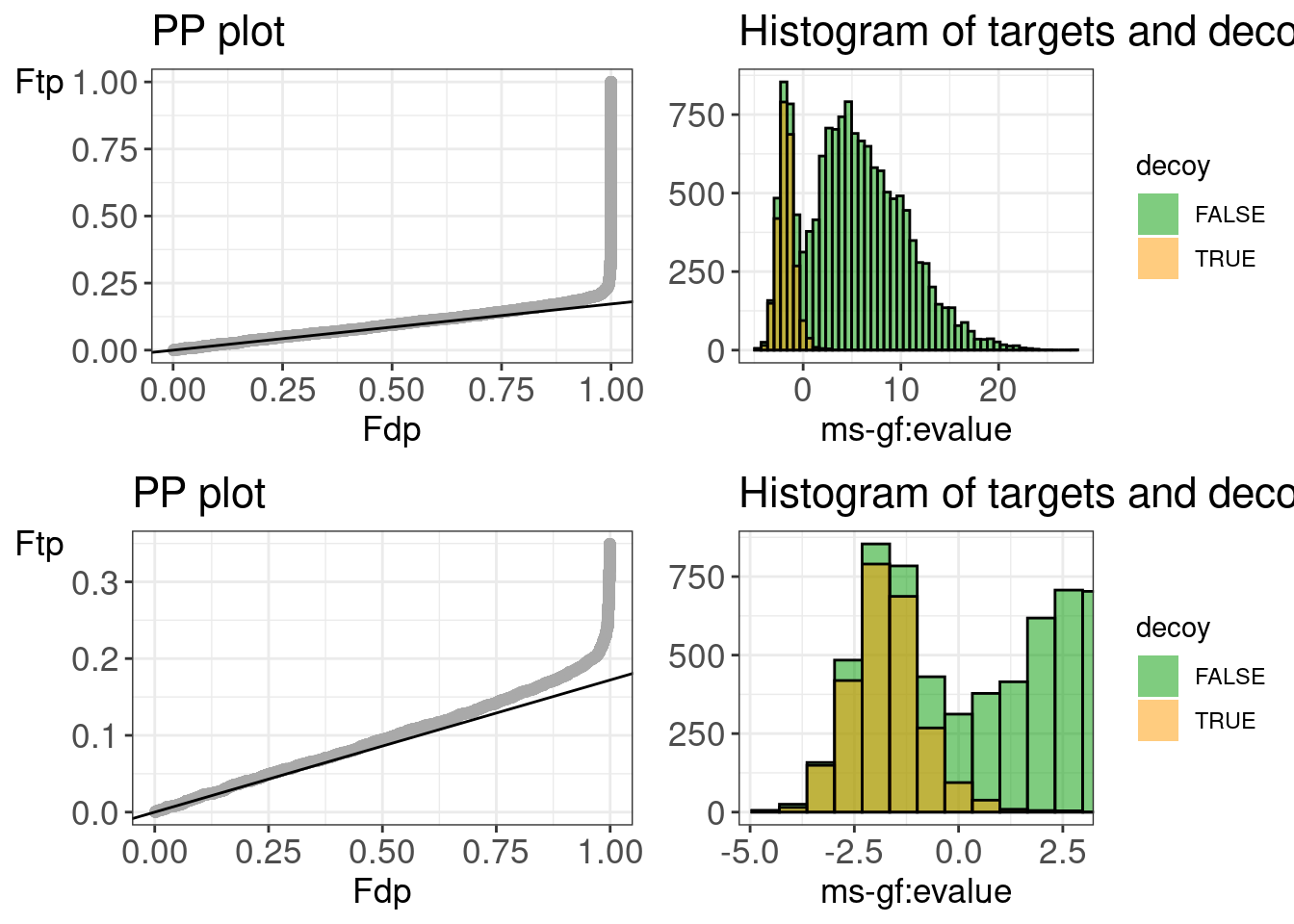
The plots show that the distribution of the MSGF+ PSM scores are nicely bimodal.
The separation between good target PSM scores and bad target PSM scores is less pronounced than for peptide shaker. So it is beneficial to include the other engines with peptideshaker.
We do not see deviations from the target decoy assumptions.
LS0tCnRpdGxlOiAiRXhhbXBsZTogTVNHRisgc2VhcmNoIG9uIFN3aXNzLVByb3QiCmF1dGhvcjogCiAgLSBuYW1lOiBMaWV2ZW4gQ2xlbWVudAogICAgYWZmaWxpYXRpb246CiAgICAtIEdoZW50IFVuaXZlcnNpdHkKb3V0cHV0OiAKICAgIGh0bWxfZG9jdW1lbnQ6CiAgICAgIGNvZGVfZG93bmxvYWQ6IHRydWUKICAgICAgdGhlbWU6IGZsYXRseQogICAgICB0b2M6IHRydWUKICAgICAgdG9jX2Zsb2F0OiB0cnVlCiAgICAgIGhpZ2hsaWdodDogdGFuZ28KICAgICAgbnVtYmVyX3NlY3Rpb25zOiB0cnVlCmxpbmtjb2xvcjogYmx1ZQp1cmxjb2xvcjogYmx1ZQpjaXRlY29sb3I6IGJsdWUKLS0tCgojIExvYWQgTGlicmFyaWVzIAoKYGBge3J9CmxpYnJhcnkoVGFyZ2V0RGVjb3kpCmxpYnJhcnkoUkN1cmwpCmxpYnJhcnkobXpJRCkKYGBgCgojIERvd25sb2FkIGRhdGEgaW4gd29ya2luZyBkaXJlY3RvcnkKCmBgYHtyfQpkb3dubG9hZC5maWxlKCAKICB1cmwgPSAiaHR0cHM6Ly9yYXcuZ2l0aHVidXNlcmNvbnRlbnQuY29tL3N0YXRPbWljcy9QREEyMkdUUEIvZGF0YS9pZGVudGlmaWNhdGlvbi9weXJvY29jY3VzTVNHRiUyQi5temlkIiwKICBkZXN0ZmlsZSA9ICJweXJvY29jY3VzTVNHRisubXppZCIKICApCmBgYAoKIyBMb2FkIERhdGEgaW4gUgoKYGBge3J9CnBhdGgyRmlsZSA8LSAicHlyb2NvY2N1c01TR0YrLm16aWQiCm1zZ2YgPC0gbXpJRChwYXRoMkZpbGUpCmBgYAoKIyBMYXVuY2ggdGhlIFNoaW55IEdhZGdldCAKCkV4cGxvcmUgdGhlIHJlc3VsdHMgZm9yIHNlYXJjaCBlbmluZ2Ugc2NvcmVzIHRvIGZpbmQgY29ycmVjdCBuYW1lcyBvZiBzZWFyY2ggZW5naW5lIHNjb3JlcyBpbiB0aGUgbXpJRC4KCmBgYHtyIGV2YWw9RkFMU0V9CmV2YWxUYXJnZXREZWNveXMobXNnZikKYGBgCgojIEV2YWx1YXRlIHRhcmdldCBkZWNveSBhc3N1bXB0aW9ucyAKCgpgYGB7cn0KZXZhbFRhcmdldERlY295cyhtc2dmLCJpc2RlY295IiwibXMtZ2Y6ZXZhbHVlIikKYGBgCgotIFRoZSBwbG90cyBzaG93IHRoYXQKdGhlIGRpc3RyaWJ1dGlvbiBvZiB0aGUgTVNHRisgUFNNIHNjb3JlcyBhcmUgbmljZWx5IGJpbW9kYWwuIAoKLSBUaGUgc2VwYXJhdGlvbiBiZXR3ZWVuIGdvb2QgdGFyZ2V0IFBTTSBzY29yZXMgYW5kIGJhZCB0YXJnZXQgUFNNIHNjb3JlcyBpcyBsZXNzIHByb25vdW5jZWQgdGhhbiBmb3IgcGVwdGlkZSBzaGFrZXIuIFNvIGl0IGlzIGJlbmVmaWNpYWwgdG8gaW5jbHVkZSB0aGUgb3RoZXIgZW5naW5lcyB3aXRoIHBlcHRpZGVzaGFrZXIuIAoKLSBXZSBkbyBub3Qgc2VlIGRldmlhdGlvbnMgZnJvbSB0aGUgdGFyZ2V0IGRlY295IGFzc3VtcHRpb25zLiAKCg==