Statistical Methods for Quantitative MS-based Proteomics: Part I. Preprocessing
Lieven Clement
statOmics, Ghent University
This is part of the online course Proteomics Data Analysis (PDA)
Outline
Introduction
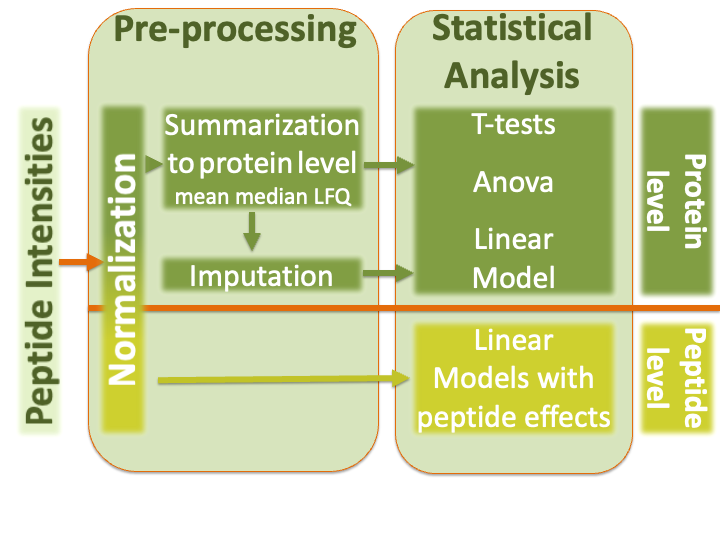
Preprocessing
- Log-transformation
- Filtering
- Normalization
- Summarization
Note, that the R-code is included for learners who are aiming to develop R/markdown scripts to automate their quantitative proteomics data analyses. According to the target audience of the course we either work with a graphical user interface (GUI) in a R/shiny App msqrob2gui (e.g. Proteomics Bioinformatics course of the EBI and the Proteomics Data Analysis course at the Gulbenkian institute) or with R/markdowns scripts (e.g. Bioinformatics Summer School at UCLouvain or the Statistical Genomics Course at Ghent University).
1 Intro: Challenges in Label-Free Quantitative Proteomics
1.1 MS-based workflow
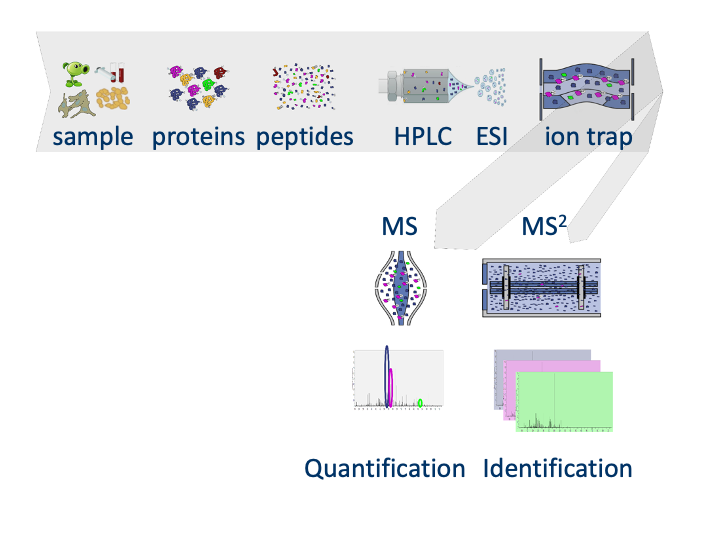
Peptide Characteristics
- Modifications
- Ionisation Efficiency: huge variability
- Identification
- Misidentification \(\rightarrow\) outliers
- MS\(^2\) selection on peptide abundance
- Context depending missingness
- Non-random missingness
\(\rightarrow\) Unbalanced pepide identifications across samples and messy data
1.2 Level of quantification
- MS-based proteomics returns peptides: pieces of proteins
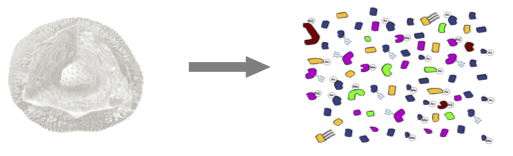
- Quantification commonly required on the protein level
1.3 Label-free Quantitative Proteomics Data Analysis Workflows
1.4 CPTAC Spike-in Study
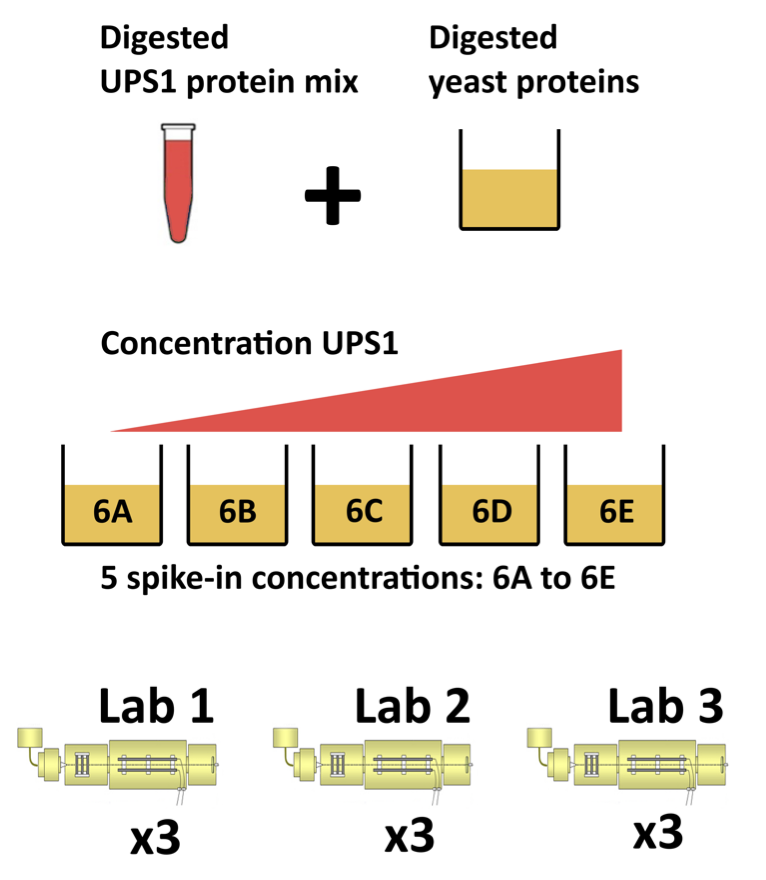
Same trypsin-digested yeast proteome background in each sample
Trypsin-digested Sigma UPS1 standard: 48 different human proteins spiked in at 5 different concentrations (treatment A-E)
Samples repeatedly run on different instruments in different labs
After MaxQuant search with match between runs option
- 41% of all proteins are quantified in all samples
- 6.6% of all peptides are quantified in all samples
\(\rightarrow\) vast amount of missingness
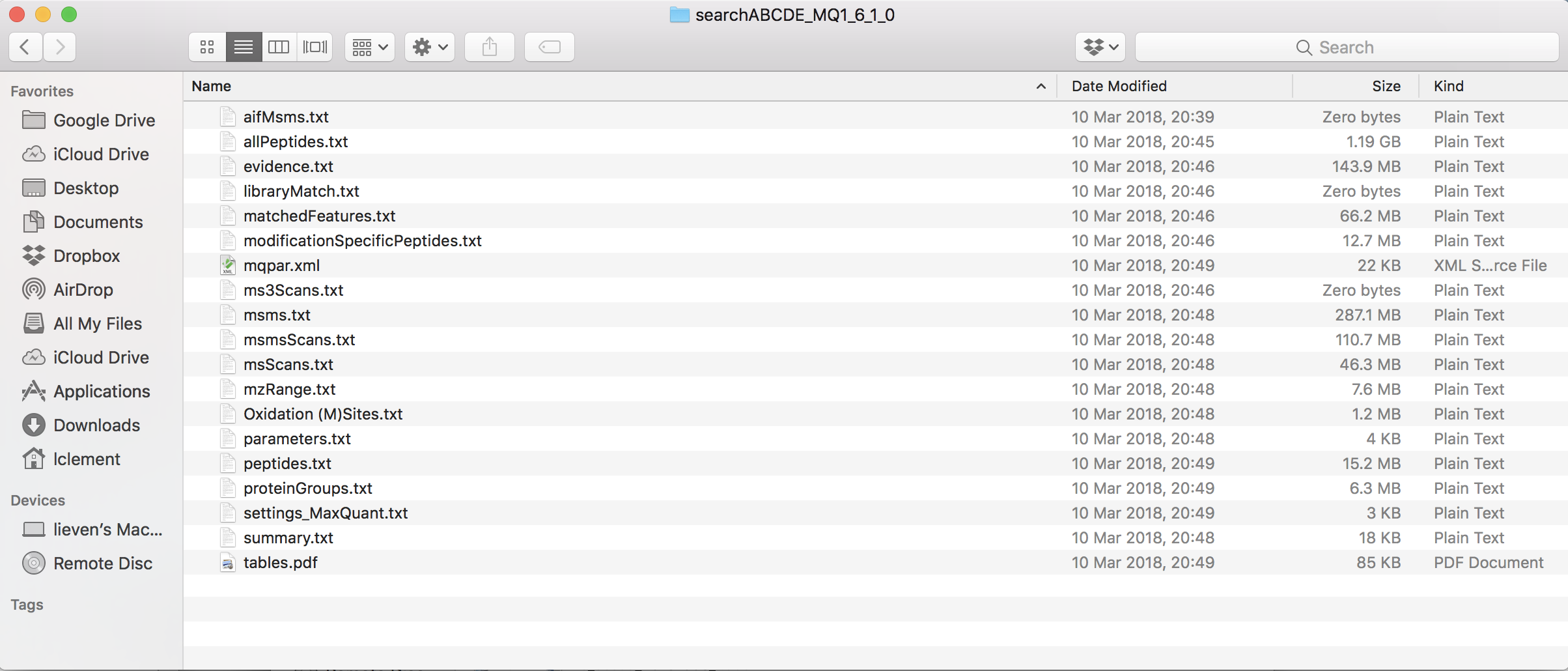
1.5 Maxquant output
2 Import the data in R
2.1 Data infrastructure
Click to see background on data infrastructure used in R to store proteomics data
- We use the
QFeaturespackage that provides the infrastructure to- store,
- process,
- manipulate and
- analyse quantitative data/features from mass spectrometry experiments.
- store,
- It is based on the
SummarizedExperimentandMultiAssayExperimentclasses.
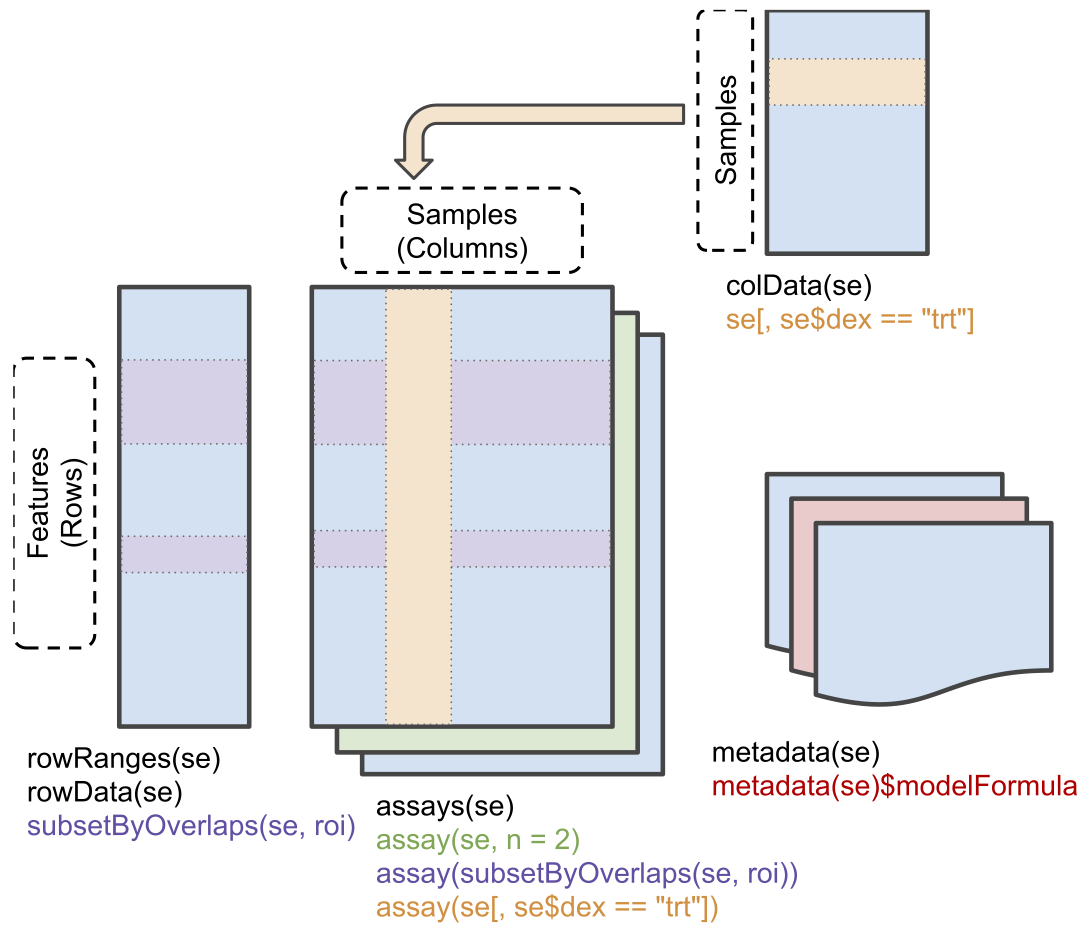
Conceptual representation of a SummarizedExperiment object.
Assays contain information on the measured omics features (rows) for
different samples (columns). The rowData contains
information on the omics features, the colData contains
information on the samples, i.e. experimental design etc.
Assays in a QFeatures object have a hierarchical relation:
- proteins are composed of peptides,
- themselves produced by spectra
- relations between assays are tracked and recorded throughout data processing
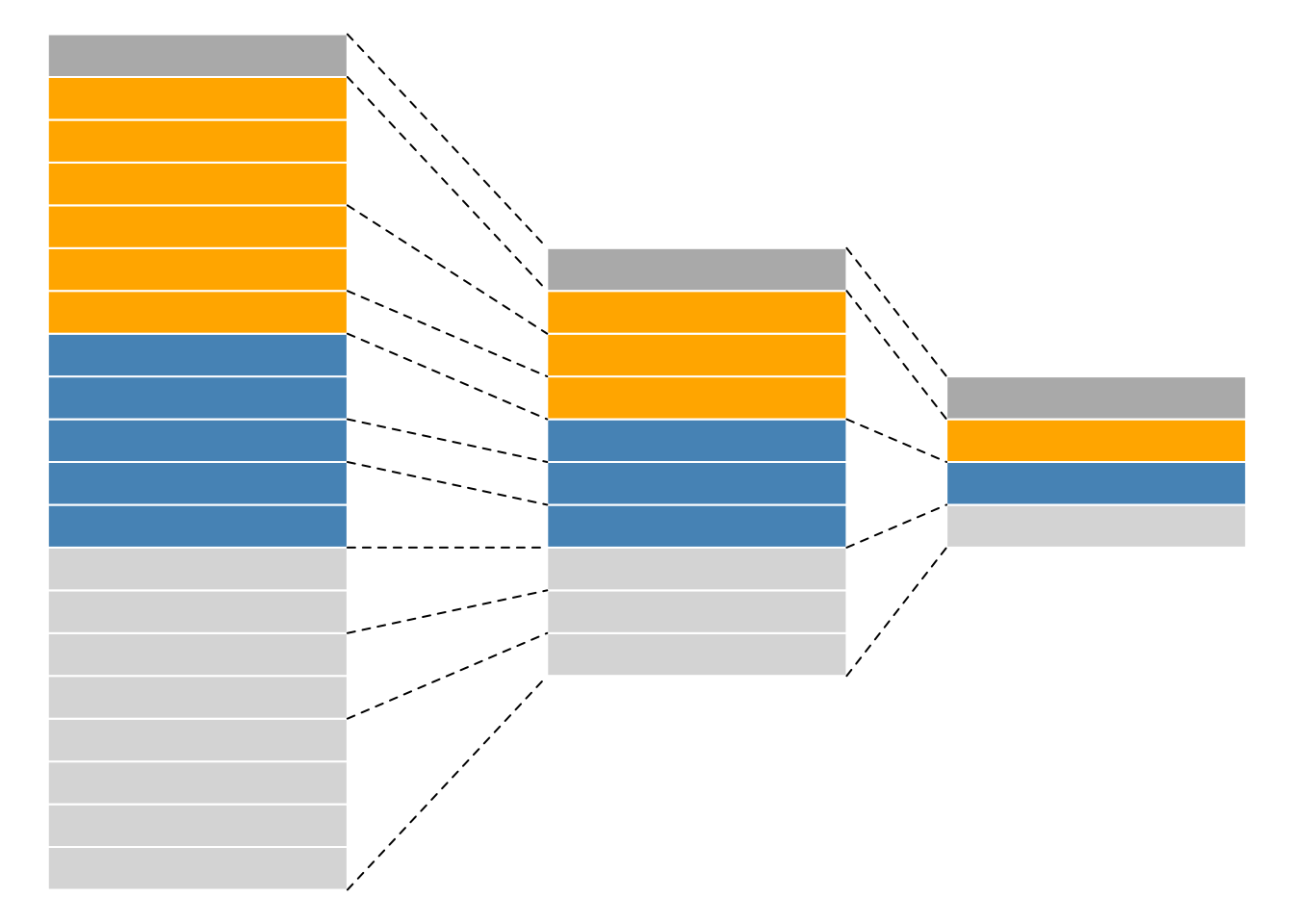
Conceptual representation of a QFeatures object and the
aggregative relation between different assays.
2.2 Import data in R
2.2.1 Load libraries
2.2.2 Read data
Click to see background and code
- We use a peptides.txt file from MS-data quantified with maxquant that contains MS1 intensities summarized at the peptide level.
peptidesFile <- "https://raw.githubusercontent.com/statOmics/PDA22GTPB/data/quantification/fullCptacDatasSetNotForTutorial/peptides.txt"- Maxquant stores the intensity data for the different samples in columnns that start with Intensity. We can retreive the column names with the intensity data with the code below:
- Read the data and store it in QFeatures object
pe <- readQFeatures(
assayData = read.delim(peptidesFile),
fnames = 1,
quantCols = ecols,
name = "peptideRaw")## Checking arguments.## Loading data as a 'SummarizedExperiment' object.## Formatting sample annotations (colData).## Formatting data as a 'QFeatures' object.## Setting assay rownames.2.2.3 Explore object
Click to see background and code
- The rowData contains information on the features (peptides) in the assay. E.g. Sequence, protein, …
## DataFrame with 11466 rows and 143 columns
## Sequence N.term.cleavage.window C.term.cleavage.window
## <character> <character> <character>
## AAAAGAGGAGDSGDAVTK AAAAGAGGAG... EHQHDEQKAA... DSGDAVTKIG...
## AAAALAGGK AAAALAGGK QQLSKAAKAA... AAALAGGKKS...
## AAAALAGGKK AAAALAGGKK QQLSKAAKAA... AALAGGKKSK...
## AAADALSDLEIK AAADALSDLE... MPKETPSKAA... ALSDLEIKDS...
## AAADALSDLEIKDSK AAADALSDLE... MPKETPSKAA... DLEIKDSKSN...
## ... ... ... ...
## YYSIYDLGNNAVGLAK YYSIYDLGNN... VGDAFLRKYY... NNAVGLAKAI...
## YYTFNGPNYNENETIR YYTFNGPNYN... FKDGSYPKYY... YNENETIRHI...
## YYTITEVATR YYTITEVATR QEWDINERYY... TITEVATRAK...
## YYTVFDRDNNR YYTVFDRDNN... LGDVFIGRYY... VFDRDNNRVG...
## YYTVFDRDNNRVGFAEAAR YYTVFDRDNN... LGDVFIGRYY... VGFAEAARL_...
## Amino.acid.before First.amino.acid Second.amino.acid
## <character> <character> <character>
## AAAAGAGGAGDSGDAVTK K A A
## AAAALAGGK K A A
## AAAALAGGKK K A A
## AAADALSDLEIK K A A
## AAADALSDLEIKDSK K A A
## ... ... ... ...
## YYSIYDLGNNAVGLAK K Y Y
## YYTFNGPNYNENETIR K Y Y
## YYTITEVATR R Y Y
## YYTVFDRDNNR R Y Y
## YYTVFDRDNNRVGFAEAAR R Y Y
## Second.last.amino.acid Last.amino.acid Amino.acid.after
## <character> <character> <character>
## AAAAGAGGAGDSGDAVTK T K I
## AAAALAGGK G K K
## AAAALAGGKK K K S
## AAADALSDLEIK I K D
## AAADALSDLEIKDSK S K S
## ... ... ... ...
## YYSIYDLGNNAVGLAK A K A
## YYTFNGPNYNENETIR I R H
## YYTITEVATR T R A
## YYTVFDRDNNR N R V
## YYTVFDRDNNRVGFAEAAR A R L
## A.Count R.Count N.Count D.Count C.Count Q.Count
## <integer> <integer> <integer> <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 7 0 0 2 0 0
## AAAALAGGK 5 0 0 0 0 0
## AAAALAGGKK 5 0 0 0 0 0
## AAADALSDLEIK 4 0 0 2 0 0
## AAADALSDLEIKDSK 4 0 0 3 0 0
## ... ... ... ... ... ... ...
## YYSIYDLGNNAVGLAK 2 0 2 1 0 0
## YYTFNGPNYNENETIR 0 1 4 0 0 0
## YYTITEVATR 1 1 0 0 0 0
## YYTVFDRDNNR 0 2 2 2 0 0
## YYTVFDRDNNRVGFAEAAR 3 3 2 2 0 0
## E.Count G.Count H.Count I.Count L.Count K.Count
## <integer> <integer> <integer> <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 0 5 0 0 0 1
## AAAALAGGK 0 2 0 0 1 1
## AAAALAGGKK 0 2 0 0 1 2
## AAADALSDLEIK 1 0 0 1 2 1
## AAADALSDLEIKDSK 1 0 0 1 2 2
## ... ... ... ... ... ... ...
## YYSIYDLGNNAVGLAK 0 2 0 1 2 1
## YYTFNGPNYNENETIR 2 1 0 1 0 0
## YYTITEVATR 1 0 0 1 0 0
## YYTVFDRDNNR 0 0 0 0 0 0
## YYTVFDRDNNRVGFAEAAR 1 1 0 0 0 0
## M.Count F.Count P.Count S.Count T.Count W.Count
## <integer> <integer> <integer> <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 0 0 0 1 1 0
## AAAALAGGK 0 0 0 0 0 0
## AAAALAGGKK 0 0 0 0 0 0
## AAADALSDLEIK 0 0 0 1 0 0
## AAADALSDLEIKDSK 0 0 0 2 0 0
## ... ... ... ... ... ... ...
## YYSIYDLGNNAVGLAK 0 0 0 1 0 0
## YYTFNGPNYNENETIR 0 1 1 0 2 0
## YYTITEVATR 0 0 0 0 3 0
## YYTVFDRDNNR 0 1 0 0 1 0
## YYTVFDRDNNRVGFAEAAR 0 2 0 0 1 0
## Y.Count V.Count U.Count Length Missed.cleavages
## <integer> <integer> <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 0 1 0 18 0
## AAAALAGGK 0 0 0 9 0
## AAAALAGGKK 0 0 0 10 1
## AAADALSDLEIK 0 0 0 12 0
## AAADALSDLEIKDSK 0 0 0 15 1
## ... ... ... ... ... ...
## YYSIYDLGNNAVGLAK 3 1 0 16 0
## YYTFNGPNYNENETIR 3 0 0 16 0
## YYTITEVATR 2 1 0 10 0
## YYTVFDRDNNR 2 1 0 11 1
## YYTVFDRDNNRVGFAEAAR 2 2 0 19 2
## Mass Proteins Leading.razor.protein
## <numeric> <character> <character>
## AAAAGAGGAGDSGDAVTK 1445.675 sp|P38915|... sp|P38915|...
## AAAALAGGK 728.418 sp|Q3E792|... sp|Q3E792|...
## AAAALAGGKK 856.513 sp|Q3E792|... sp|Q3E792|...
## AAADALSDLEIK 1215.635 sp|P09938|... sp|P09938|...
## AAADALSDLEIKDSK 1545.789 sp|P09938|... sp|P09938|...
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1759.88 sp|P07267|... sp|P07267|...
## YYTFNGPNYNENETIR 1993.88 sp|Q00955|... sp|Q00955|...
## YYTITEVATR 1215.61 sp|P38891|... sp|P38891|...
## YYTVFDRDNNR 1461.66 P07339ups|... P07339ups|...
## YYTVFDRDNNRVGFAEAAR 2263.08 P07339ups|... P07339ups|...
## Start.position End.position Unique..Groups.
## <integer> <integer> <character>
## AAAAGAGGAGDSGDAVTK 97 114 yes
## AAAALAGGK 13 21 yes
## AAAALAGGKK 13 22 yes
## AAADALSDLEIK 9 20 yes
## AAADALSDLEIKDSK 9 23 yes
## ... ... ... ...
## YYSIYDLGNNAVGLAK 388 403 yes
## YYTFNGPNYNENETIR 1275 1290 yes
## YYTITEVATR 311 320 yes
## YYTVFDRDNNR 225 235 yes
## YYTVFDRDNNRVGFAEAAR 225 243 yes
## Unique..Proteins. Charges PEP Score
## <character> <character> <numeric> <numeric>
## AAAAGAGGAGDSGDAVTK yes 2 1.1843e-05 82.942
## AAAALAGGK no 2 7.4562e-06 134.810
## AAAALAGGKK no 2 3.3094e-09 143.730
## AAADALSDLEIK yes 2 9.1593e-23 182.230
## AAADALSDLEIKDSK yes 3 1.5319e-04 73.927
## ... ... ... ... ...
## YYSIYDLGNNAVGLAK yes 2 7.7415e-37 174.240
## YYTFNGPNYNENETIR yes 2 4.2208e-21 147.750
## YYTITEVATR yes 2 1.3566e-04 109.160
## YYTVFDRDNNR yes 2 6.1425e-04 110.930
## YYTVFDRDNNRVGFAEAAR yes 3 8.9859e-04 59.728
## Identification.type.6A_1 Identification.type.6A_2
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By MS/MS
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By matchin... By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6A_3 Identification.type.6A_4
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By MS/MS
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By MS/MS
## AAADALSDLEIK By matchin... By MS/MS
## AAADALSDLEIKDSK By matchin... By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By MS/MS
## YYTFNGPNYNENETIR By matchin... By MS/MS
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6A_5 Identification.type.6A_6
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By MS/MS
## YYTFNGPNYNENETIR By MS/MS By MS/MS
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6A_7 Identification.type.6A_8
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By MS/MS
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By MS/MS By matchin...
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6A_9 Identification.type.6B_1
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By matchin...
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By MS/MS
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6B_2 Identification.type.6B_3
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By MS/MS By matchin...
## AAADALSDLEIKDSK By matchin... By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6B_4 Identification.type.6B_5
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By matchin...
## YYTFNGPNYNENETIR By MS/MS By MS/MS
## YYTITEVATR By MS/MS By MS/MS
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6B_6 Identification.type.6B_7
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By MS/MS
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By MS/MS By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6B_8 Identification.type.6B_9
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By MS/MS
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By matchin... By matchin...
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6C_1 Identification.type.6C_2
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By MS/MS
## AAADALSDLEIK By MS/MS By matchin...
## AAADALSDLEIKDSK By matchin... By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6C_3 Identification.type.6C_4
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By MS/MS
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By matchin... By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By MS/MS
## YYTFNGPNYNENETIR By matchin... By MS/MS
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6C_5 Identification.type.6C_6
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By MS/MS
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6C_7 Identification.type.6C_8
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By matchin...
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By matchin... By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By MS/MS
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6C_9 Identification.type.6D_1
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By MS/MS By matchin...
## AAAALAGGKK By MS/MS By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6D_2 Identification.type.6D_3
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By matchin... By matchin...
## AAADALSDLEIKDSK By MS/MS By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By MS/MS
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6D_4 Identification.type.6D_5
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By MS/MS By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By MS/MS
## YYTFNGPNYNENETIR By MS/MS By MS/MS
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6D_6 Identification.type.6D_7
## <character> <character>
## AAAAGAGGAGDSGDAVTK By MS/MS By matchin...
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By MS/MS
## AAADALSDLEIK By MS/MS By matchin...
## AAADALSDLEIKDSK By matchin... By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By matchin...
## YYTFNGPNYNENETIR By MS/MS By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By MS/MS
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6D_8 Identification.type.6D_9
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By MS/MS By matchin...
## YYTVFDRDNNR By MS/MS By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6E_1 Identification.type.6E_2
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6E_3 Identification.type.6E_4
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By MS/MS
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By matchin... By MS/MS
## AAADALSDLEIKDSK By MS/MS By matchin...
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By MS/MS
## YYTFNGPNYNENETIR By matchin... By MS/MS
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By matchin... By MS/MS
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6E_5 Identification.type.6E_6
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By matchin... By matchin...
## AAAALAGGKK By matchin... By matchin...
## AAADALSDLEIK By MS/MS By matchin...
## AAADALSDLEIKDSK By MS/MS By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By MS/MS By MS/MS
## YYTFNGPNYNENETIR By MS/MS By MS/MS
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By MS/MS By matchin...
## YYTVFDRDNNRVGFAEAAR By matchin... By MS/MS
## Identification.type.6E_7 Identification.type.6E_8
## <character> <character>
## AAAAGAGGAGDSGDAVTK By matchin... By matchin...
## AAAALAGGK By MS/MS By MS/MS
## AAAALAGGKK By MS/MS By MS/MS
## AAADALSDLEIK By MS/MS By MS/MS
## AAADALSDLEIKDSK By matchin... By MS/MS
## ... ... ...
## YYSIYDLGNNAVGLAK By matchin... By matchin...
## YYTFNGPNYNENETIR By matchin... By matchin...
## YYTITEVATR By matchin... By matchin...
## YYTVFDRDNNR By MS/MS By MS/MS
## YYTVFDRDNNRVGFAEAAR By matchin... By matchin...
## Identification.type.6E_9 Experiment.6A_1 Experiment.6A_2
## <character> <integer> <integer>
## AAAAGAGGAGDSGDAVTK By matchin... NA 1
## AAAALAGGK By MS/MS NA 1
## AAAALAGGKK By MS/MS NA 1
## AAADALSDLEIK By MS/MS 1 1
## AAADALSDLEIKDSK By MS/MS 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK By matchin... NA NA
## YYTFNGPNYNENETIR By MS/MS NA NA
## YYTITEVATR By matchin... 1 NA
## YYTVFDRDNNR By MS/MS NA NA
## YYTVFDRDNNRVGFAEAAR By matchin... NA NA
## Experiment.6A_3 Experiment.6A_4 Experiment.6A_5
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA 1 1
## AAAALAGGK 2 1 1
## AAAALAGGKK NA 1 NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK NA 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA 1 1
## YYTFNGPNYNENETIR NA 1 1
## YYTITEVATR 1 NA NA
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6A_6 Experiment.6A_7 Experiment.6A_8
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 1 1
## AAAALAGGK 1 2 1
## AAAALAGGKK 1 1 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 NA NA
## YYTFNGPNYNENETIR 1 1 NA
## YYTITEVATR 1 1 NA
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6A_9 Experiment.6B_1 Experiment.6B_2
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 NA NA
## AAAALAGGK 1 1 1
## AAAALAGGKK 1 NA 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 NA 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA NA NA
## YYTFNGPNYNENETIR 1 NA NA
## YYTITEVATR NA 1 1
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6B_3 Experiment.6B_4 Experiment.6B_5
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA NA 1
## AAAALAGGK 1 2 1
## AAAALAGGKK 1 1 NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK NA 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA 1 1
## YYTFNGPNYNENETIR NA 1 1
## YYTITEVATR 1 1 1
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6B_6 Experiment.6B_7 Experiment.6B_8
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 NA 1
## AAAALAGGK NA 2 1
## AAAALAGGKK NA 1 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 NA NA
## YYTFNGPNYNENETIR 1 1 NA
## YYTITEVATR 1 NA 1
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6B_9 Experiment.6C_1 Experiment.6C_2
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 NA NA
## AAAALAGGK 2 NA 1
## AAAALAGGKK 1 NA 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA NA NA
## YYTFNGPNYNENETIR NA NA NA
## YYTITEVATR NA 1 1
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6C_3 Experiment.6C_4 Experiment.6C_5
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA 1 1
## AAAALAGGK 2 2 NA
## AAAALAGGKK NA 1 NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA 1 1
## YYTFNGPNYNENETIR NA 1 1
## YYTITEVATR 1 1 NA
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6C_6 Experiment.6C_7 Experiment.6C_8
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 1 1
## AAAALAGGK NA 2 1
## AAAALAGGKK NA 1 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 NA NA
## YYTFNGPNYNENETIR 1 1 1
## YYTITEVATR 1 NA 1
## YYTVFDRDNNR 1 NA 1
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6C_9 Experiment.6D_1 Experiment.6D_2
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 NA NA
## AAAALAGGK 1 NA 1
## AAAALAGGKK 1 NA NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA NA NA
## YYTFNGPNYNENETIR 1 NA NA
## YYTITEVATR 1 NA 1
## YYTVFDRDNNR NA NA NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6D_3 Experiment.6D_4 Experiment.6D_5
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA 1 1
## AAAALAGGK 1 1 1
## AAAALAGGKK NA 1 NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA 1 1
## YYTFNGPNYNENETIR NA 1 1
## YYTITEVATR 1 1 1
## YYTVFDRDNNR NA 1 1
## YYTVFDRDNNRVGFAEAAR NA 1 NA
## Experiment.6D_6 Experiment.6D_7 Experiment.6D_8
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 1 NA
## AAAALAGGK NA 2 1
## AAAALAGGKK NA 1 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 1 NA
## YYTFNGPNYNENETIR 1 1 1
## YYTITEVATR 1 NA 1
## YYTVFDRDNNR 1 1 1
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6D_9 Experiment.6E_1 Experiment.6E_2
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA NA 1
## AAAALAGGK 2 NA 1
## AAAALAGGKK 1 NA NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK NA NA NA
## YYTFNGPNYNENETIR 1 NA NA
## YYTITEVATR NA NA 1
## YYTVFDRDNNR 1 1 NA
## YYTVFDRDNNRVGFAEAAR NA NA NA
## Experiment.6E_3 Experiment.6E_4 Experiment.6E_5
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK NA NA 1
## AAAALAGGK 2 2 1
## AAAALAGGKK NA 1 NA
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 1 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 1 1
## YYTFNGPNYNENETIR NA 1 1
## YYTITEVATR 1 1 1
## YYTVFDRDNNR 1 1 1
## YYTVFDRDNNRVGFAEAAR NA 1 1
## Experiment.6E_6 Experiment.6E_7 Experiment.6E_8
## <integer> <integer> <integer>
## AAAAGAGGAGDSGDAVTK 1 NA NA
## AAAALAGGK NA 2 2
## AAAALAGGKK NA 1 1
## AAADALSDLEIK 1 1 1
## AAADALSDLEIKDSK 1 NA 1
## ... ... ... ...
## YYSIYDLGNNAVGLAK 1 NA NA
## YYTFNGPNYNENETIR 1 1 1
## YYTITEVATR NA NA NA
## YYTVFDRDNNR 1 1 1
## YYTVFDRDNNRVGFAEAAR 1 1 1
## Experiment.6E_9 Intensity Reverse Potential.contaminant
## <integer> <numeric> <character> <character>
## AAAAGAGGAGDSGDAVTK NA 1190800
## AAAALAGGK 1 280990000
## AAAALAGGKK 1 33360000
## AAADALSDLEIK 1 54622000
## AAADALSDLEIKDSK 1 18910000
## ... ... ... ... ...
## YYSIYDLGNNAVGLAK NA 2145900
## YYTFNGPNYNENETIR 1 5608800
## YYTITEVATR NA 13034000
## YYTVFDRDNNR 1 8702500
## YYTVFDRDNNRVGFAEAAR 1 2391100
## id Protein.group.IDs Mod..peptide.IDs Evidence.IDs
## <integer> <character> <character> <character>
## AAAAGAGGAGDSGDAVTK 0 859 0 0;1;2;3;4;...
## AAAALAGGK 1 230 1 24;25;26;2...
## AAAALAGGKK 2 230 2 74;75;76;7...
## AAADALSDLEIK 3 229 3 99;100;101...
## AAADALSDLEIKDSK 4 229 4 144;145;14...
## ... ... ... ... ...
## YYSIYDLGNNAVGLAK 11461 196 12240 331367;331...
## YYTFNGPNYNENETIR 11462 1254 12241 331384;331...
## YYTITEVATR 11463 854 12242 331411;331...
## YYTVFDRDNNR 11464 34 12243 331439;331...
## YYTVFDRDNNRVGFAEAAR 11465 34 12244 331455;331...
## MS.MS.IDs Best.MS.MS Oxidation..M..site.IDs MS.MS.Count
## <character> <integer> <character> <integer>
## AAAAGAGGAGDSGDAVTK 0;1;2;3;4;... 0 10
## AAAALAGGK 10;11;12;1... 21 18
## AAAALAGGKK 30;31;32;3... 31 21
## AAADALSDLEIK 51;52;53;5... 72 29
## AAADALSDLEIKDSK 85;86;87;8... 94 32
## ... ... ... ... ...
## YYSIYDLGNNAVGLAK 169138;169... 169147 13
## YYTFNGPNYNENETIR 169151;169... 169159 14
## YYTITEVATR 169165;169... 169173 12
## YYTVFDRDNNR 169177;169... 169180 7
## YYTVFDRDNNRVGFAEAAR 169184 169184 1- The colData contains information on the samples
## DataFrame with 45 rows and 0 columns- No information is stored yet on the design.
## CharacterList of length 1
## [["peptideRaw"]] Intensity.6A_1 Intensity.6A_2 ... Intensity.6E_9Note, that the sample names include the spike-in condition.
They also end on a number.
- 1-3 is from lab 1,
- 4-6 from lab 2 and
- 7-9 from lab 3.
We update the colData with information on the design
colData(pe)$lab <- rep(rep(paste0("lab",1:3),each=3),5) %>% as.factor
colData(pe)$condition <- pe[["peptideRaw"]] %>% colnames %>% substr(12,12) %>% as.factor
colData(pe)$spikeConcentration <- rep(c(A = 0.25, B = 0.74, C = 2.22, D = 6.67, E = 20),each = 9)- We explore the colData again
## DataFrame with 45 rows and 3 columns
## lab condition spikeConcentration
## <factor> <factor> <numeric>
## Intensity.6A_1 lab1 A 0.25
## Intensity.6A_2 lab1 A 0.25
## Intensity.6A_3 lab1 A 0.25
## Intensity.6A_4 lab2 A 0.25
## Intensity.6A_5 lab2 A 0.25
## ... ... ... ...
## Intensity.6E_5 lab2 E 20
## Intensity.6E_6 lab2 E 20
## Intensity.6E_7 lab3 E 20
## Intensity.6E_8 lab3 E 20
## Intensity.6E_9 lab3 E 203 Preprocessing
3.1 Log-transformation
3.1.1 Explore the data with plots
Peptide AALEELVK from spiked-in UPS protein P12081. We only show data from lab1.
Click to see code to make plot
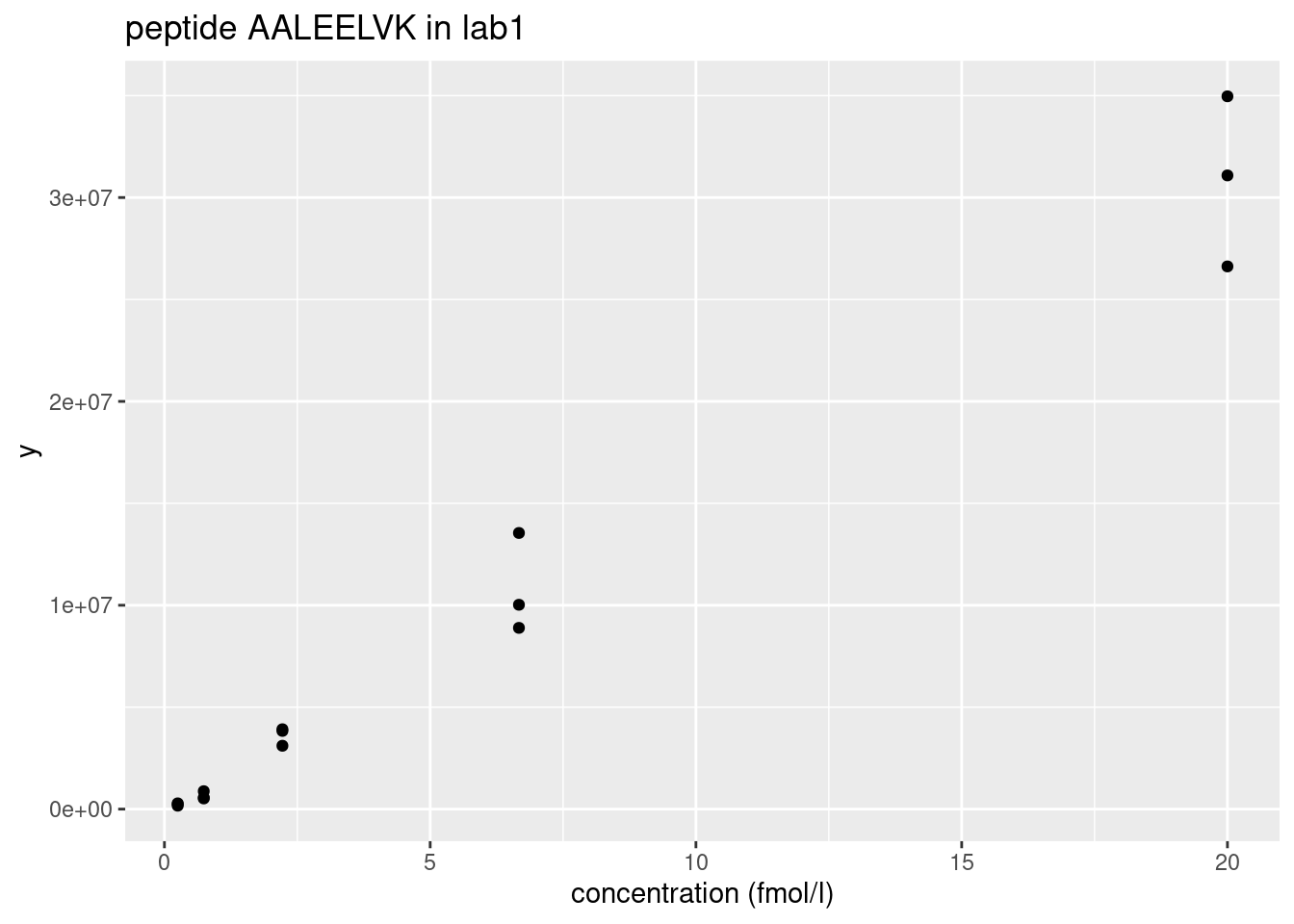
- Variance increases with the mean \(\rightarrow\) Multiplicative error structure
Click to see code to make plot
plotLog <- data.frame(concentration = colData(subset)$spikeConcentration,
y = assay(subset[["peptideRaw"]]) %>% c
) %>%
ggplot(aes(concentration, y)) +
geom_point() +
scale_x_continuous(trans='log2') +
scale_y_continuous(trans='log2') +
xlab("concentration (fmol/l)") +
ggtitle("peptide AALEELVK in lab1 with axes on log scale")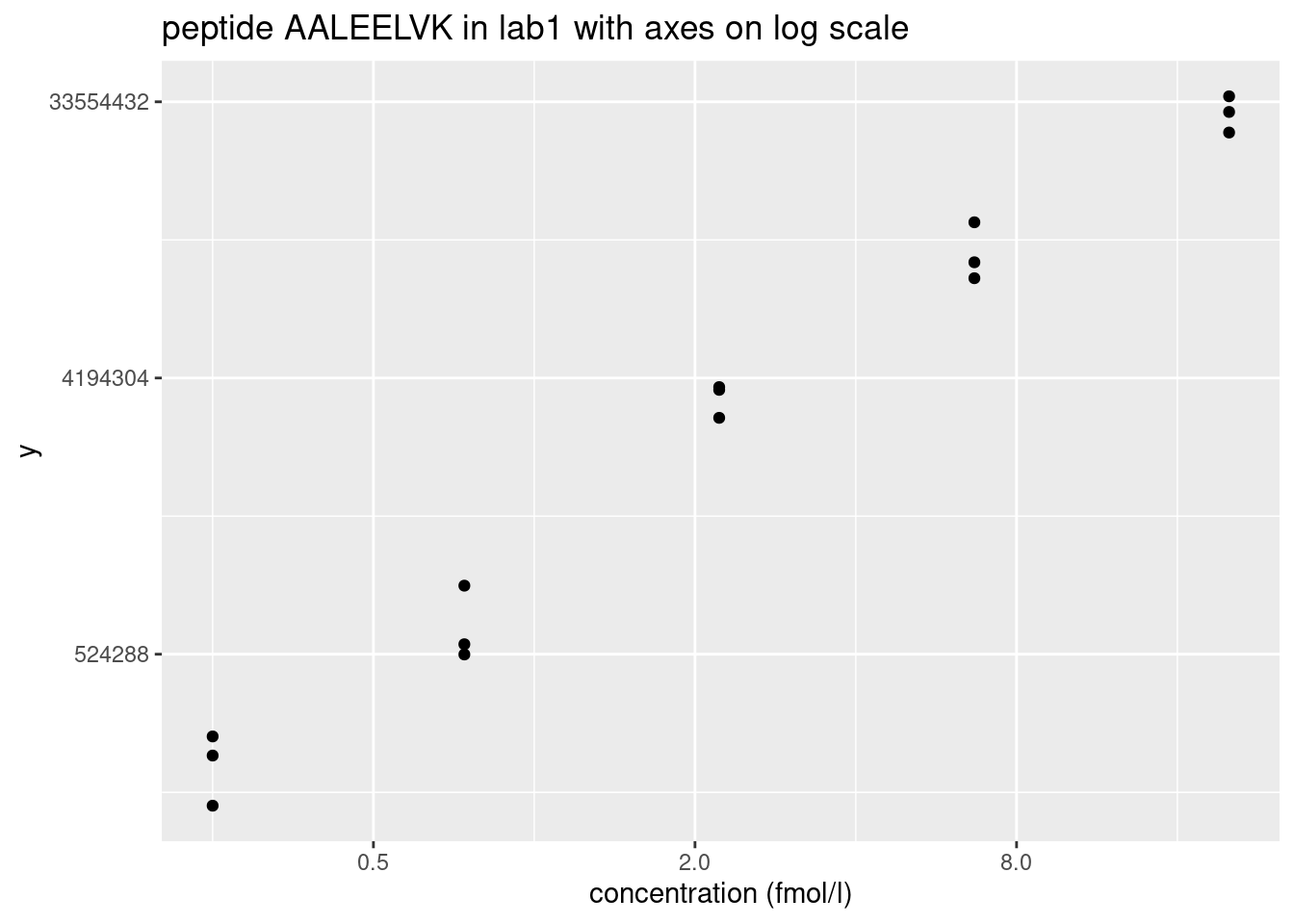
- Data seems to be homoscedastic on log-scale \(\rightarrow\) log transformation of the intensity data
- In quantitative proteomics analysis on \(\log_2\)
\(\rightarrow\) Differences on a \(\log_2\) scale: \(\log_2\) fold changes
\[ \log_2 B - \log_2 A = \log_2 \frac{B}{A} = \log FC_\text{B - A} \] \[ \begin{array} {l} log_2 FC = 1 \rightarrow FC = 2^1 =2\\ log_2 FC = 2 \rightarrow FC = 2^2 = 4\\ \end{array} \]
3.1.2 log-transformation of the data
Click to see code to log-transfrom the data
- We calculate how many non zero intensities we have for each peptide and this can be useful for filtering.
- Peptides with zero intensities are missing peptides and should be
represent with a
NAvalue rather than0.
- Logtransform data with base 2
3.2 Filtering
- Reverse sequences
- Only identified by modification site (only modified peptides detected)
- Razor peptides: non-unique peptides assigned to the protein group with the most other peptides
- Contaminants
- Peptides few identifications
- Proteins that are only identified with one or a few peptides
Filtering does not induce bias if the criterion is independent from the downstream data analysis!
Click to see code to filter the data
- Handling overlapping protein groups
In our approach a peptide can map to multiple proteins, as long as there is none of these proteins present in a smaller subgroup.
pe <- filterFeatures(pe, ~ Proteins %in% smallestUniqueGroups(rowData(pe[["peptideLog"]])$Proteins))## 'Proteins' found in 2 out of 2 assay(s).- Remove reverse sequences (decoys) and contaminants
We now remove the contaminants, peptides that map to decoy sequences, and proteins which were only identified by peptides with modifications.
## 'Reverse' found in 2 out of 2 assay(s).## 'Potential.contaminant' found in 2 out of 2 assay(s).- Drop peptides that were only identified in one sample
We keep peptides that were observed at last twice.
## 'nNonZero' found in 2 out of 2 assay(s).## [1] 104783.3 Normalization
Click to see code to make plot
densityConditionD <- pe[["peptideLog"]][,colData(pe)$condition=="D"] %>%
assay %>%
as.data.frame() %>%
gather(sample, intensity) %>%
mutate(lab = colData(pe)[sample,"lab"]) %>%
ggplot(aes(x=intensity,group=sample,color=lab)) +
geom_density() +
ggtitle("condition D")
densityLab2 <- pe[["peptideLog"]][,colData(pe)$lab=="lab2"] %>%
assay %>%
as.data.frame() %>%
gather(sample, intensity) %>%
mutate(condition = colData(pe)[sample,"condition"]) %>%
ggplot(aes(x=intensity,group=sample,color=condition)) +
geom_density() +
ggtitle("lab2")## Warning: Removed 39179 rows containing non-finite outside the scale range
## (`stat_density()`).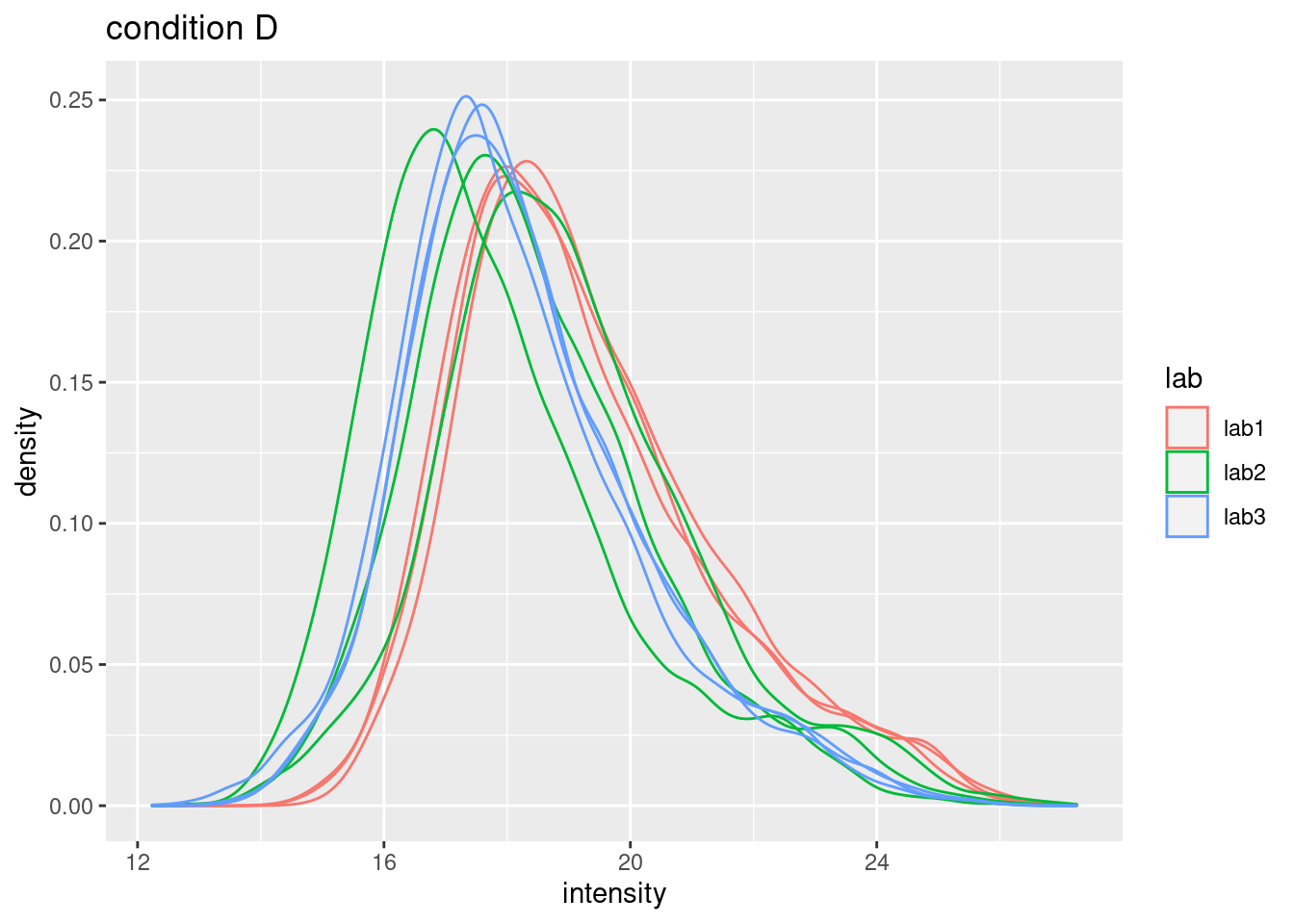
## Warning: Removed 44480 rows containing non-finite outside the scale range
## (`stat_density()`).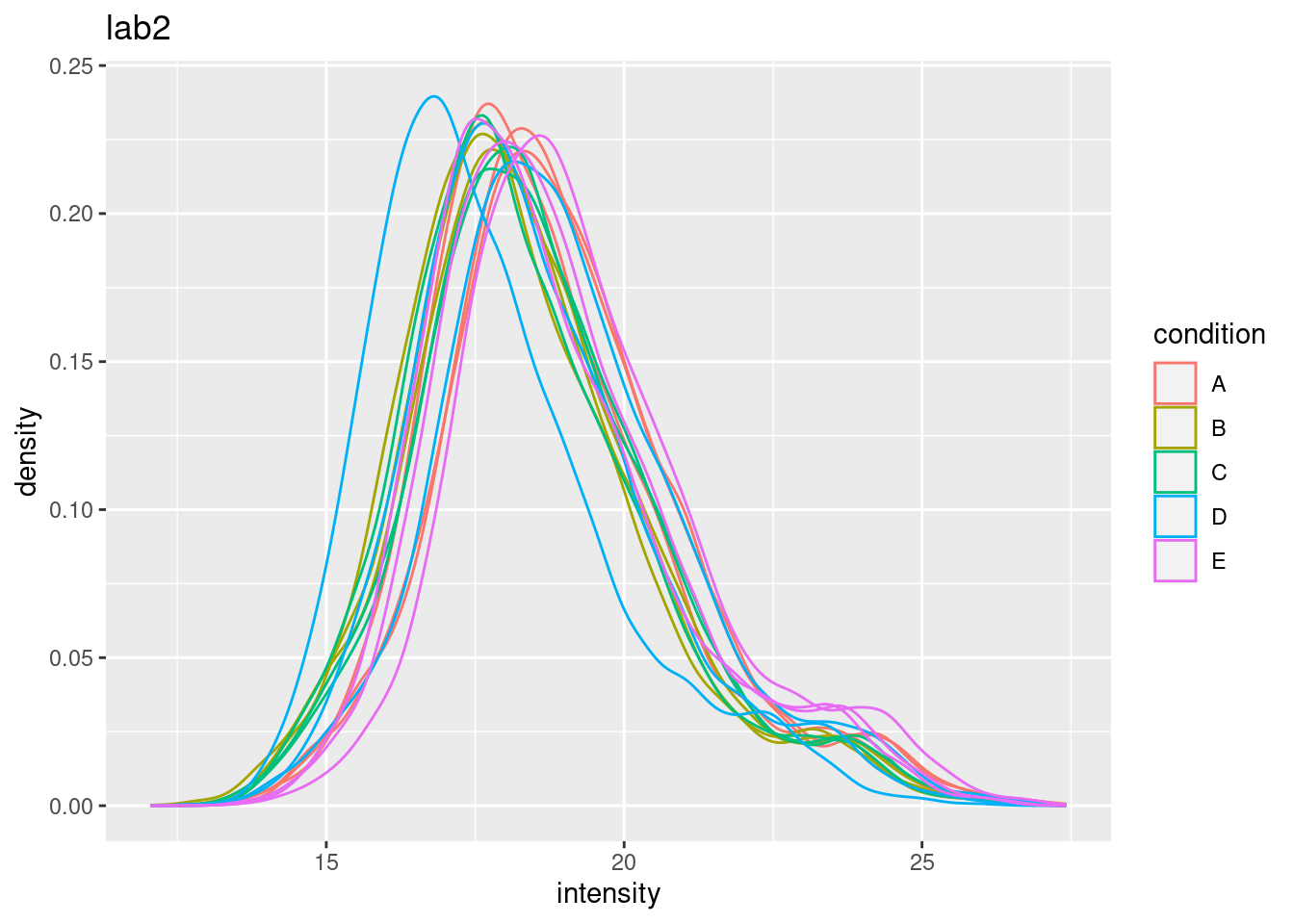 - Even in very clean synthetic dataset (same background, only 48 UPS
proteins can be different) the marginal peptide intensity distribution
across samples can be quite distinct
- Even in very clean synthetic dataset (same background, only 48 UPS
proteins can be different) the marginal peptide intensity distribution
across samples can be quite distinct
- Considerable effects between and within labs for replicate samples
- Considerable effects between samples with different spike-in concentration
\(\rightarrow\) Normalization is needed
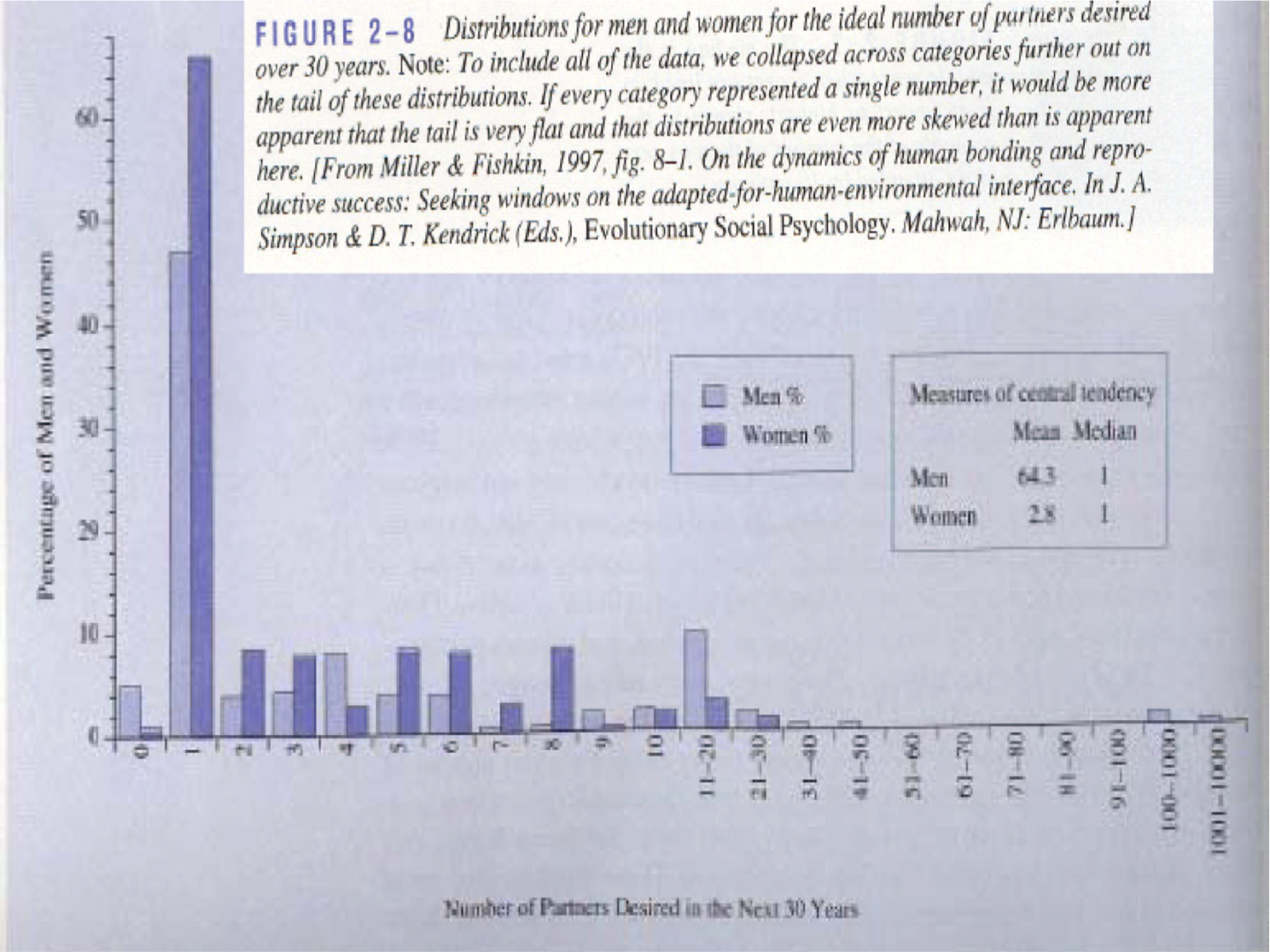
3.3.1 Mean or median?
- Miller and Fishkin (1997) reported that over a period of 30 years males would like to have on average 64.3 partners and females 2.8.
- Miller and Fishkin (1997) reported that the median number of partners someone would like to have over a period of 30 years males is 1 for both males and females.
Mean is very sensitive to outliers!
3.3.2 Normalization of the data by median centering
\[y_{ip}^\text{norm} = y_{ip} - \hat\mu_i\] with \(\hat\mu_i\) the median intensity over all observed peptides in sample \(i\).
3.3.3 Plots of normalized data
Click to see code to make plot
densityConditionDNorm <- pe[["peptideNorm"]][,colData(pe)$condition=="D"] %>%
assay %>%
as.data.frame() %>%
gather(sample, intensity) %>%
mutate(lab = colData(pe)[sample,"lab"]) %>%
ggplot(aes(x=intensity,group=sample,color=lab)) +
geom_density() +
ggtitle("condition D")
densityLab2Norm <- pe[["peptideNorm"]][,colData(pe)$lab=="lab2"] %>%
assay %>%
as.data.frame() %>%
gather(sample, intensity) %>%
mutate(condition = colData(pe)[sample,"condition"]) %>%
ggplot(aes(x=intensity,group=sample,color=condition)) +
geom_density() +
ggtitle("lab2")## Warning: Removed 39179 rows containing non-finite outside the scale range
## (`stat_density()`).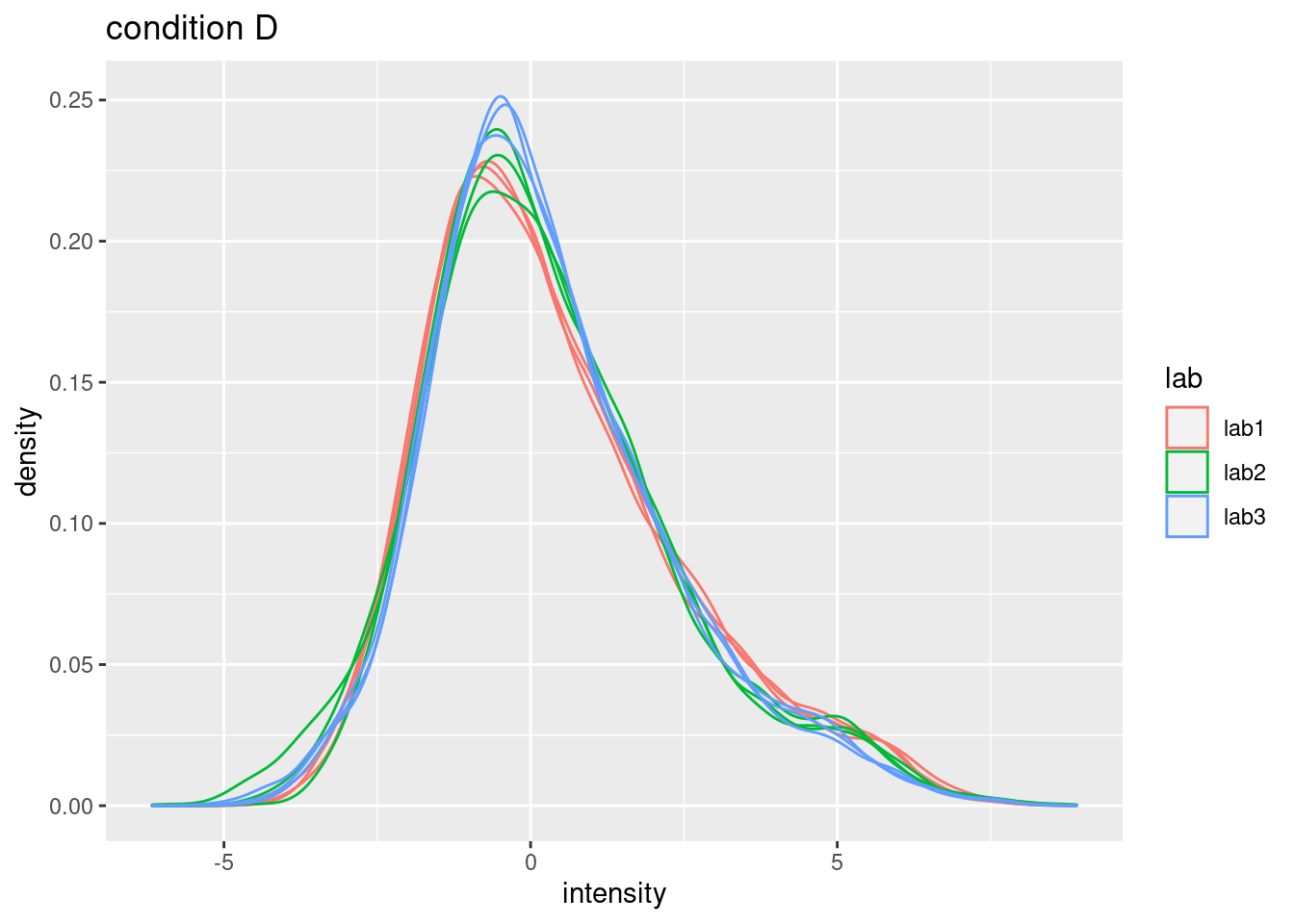
## Warning: Removed 44480 rows containing non-finite outside the scale range
## (`stat_density()`).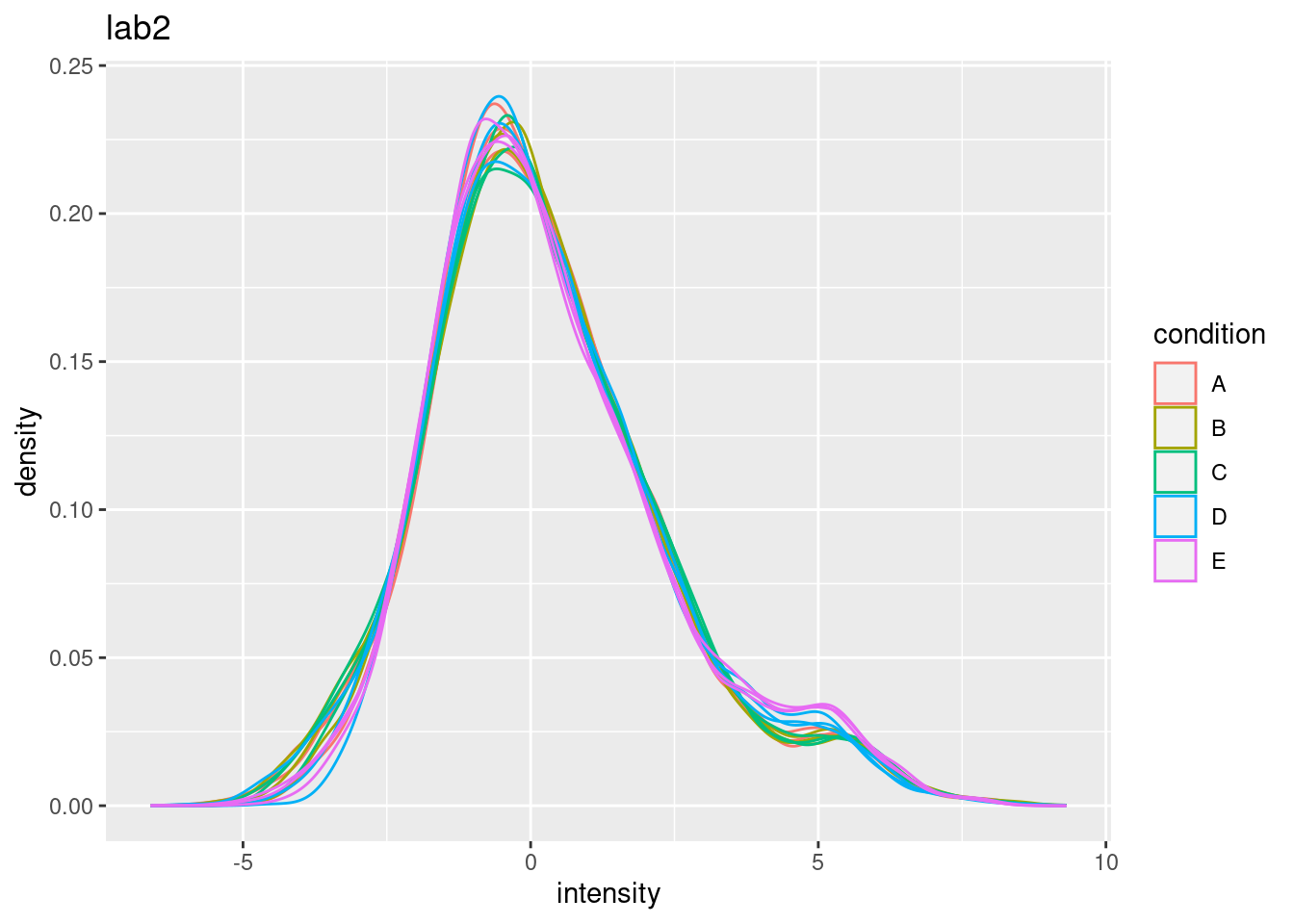
- Upon normalization the marginal distributions of the peptide intensities across samples are much more comparable
- We still see deviations
- This can be due to technical variability
- In micro-array literature, quantile normalisation is used to force the median and all other quantiles to be equal across samples
- In proteomics quantile normalisation often introduces artifacts due to a difference in missing peptides across samples
- More advanced methods should be developed for normalizing proteomics data
- If there are differences in the width of the marginal distributions of the data across samples. They can also be standardized by using a robust estimator for location and scale, i.e. \[y_{ip}^\text{norm} = \frac{y_{ip} - \mu_i}{s_i}\]
3.4 Summarization
- We illustrate summarization issues using a subset of the cptac study (Lab 2, condition A and E) for a spiked protein (UPS P12081).
Click to see code to make plot
summaryPlot <- pe[["peptideNorm"]][
rowData(pe[["peptideNorm"]])$Proteins == "P12081ups|SYHC_HUMAN_UPS",
colData(pe)$lab=="lab2"&colData(pe)$condition %in% c("A","E")] %>%
assay %>%
as.data.frame %>%
rownames_to_column(var = "peptide") %>%
gather(sample, intensity, -peptide) %>%
mutate(condition = colData(pe)[sample,"condition"]) %>%
ggplot(aes(x = peptide, y = intensity, color = sample, group = sample, label = condition), show.legend = FALSE) +
geom_line(show.legend = FALSE) +
geom_text(show.legend = FALSE) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
xlab("Peptide") +
ylab("Intensity (log2)")## Warning: Removed 10 rows containing missing values or values outside the scale range
## (`geom_line()`).## Warning: Removed 90 rows containing missing values or values outside the scale range
## (`geom_text()`).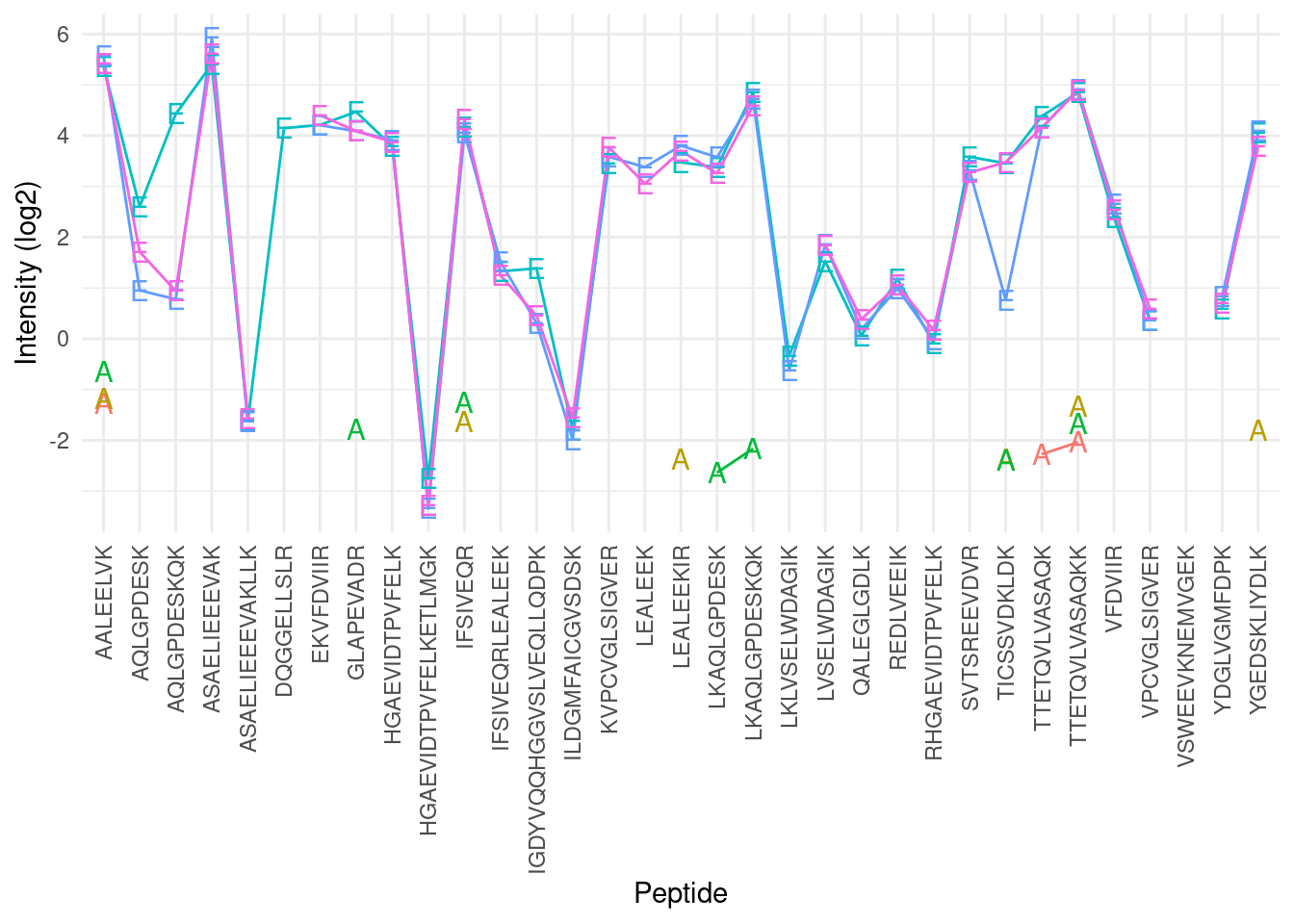
We observe:
- intensities from multiple peptides for each protein in a sample
- Strong peptide effect -Unbalanced peptide identification
- Pseudo-replication: peptide intensities from a particular protein in the same sample are correlated, i.e. they more alike than peptide intensities from a particular protein between samples.
\(\rightarrow\) Summarize all peptide intensities from the same protein in a sample into a single protein expression value
Commonly used methods are
Mean summarization \[ y_{ip}=\beta_i^\text{samp} + \epsilon_{ip} \]
Median summarization
Maxquant’s maxLFQ summarization (in protein groups file)
Model based summarization: \[ y_{ip}=\beta_i^\text{samp} + \beta_p^\text{pep} + \epsilon_{ip} \]
Click to see R-code to aggregate the data
We use the standard sumarization in aggregateFeatures, which is robust model based summarization.
## Your quantitative and row data contain missing values. Please read the
## relevant section(s) in the aggregateFeatures manual page regarding the
## effects of missing values on data aggregation.## Aggregated: 1/1Other summarization methods can be implemented by using the
fun argument in the aggregateFeatures
function.
fun = MsCoreUtils::medianPolish()to fits an additive model (two way decomposition) using Tukey’s median polish_ procedure using stats::medpolish()fun = MsCoreUtils::robustSummary()to calculate a robust aggregation using MASS::rlm() (default)fun = base::colMeans()to use the mean of each columnfun = matrixStats::colMedians()to use the median of each columnfun = base::colSums()to use the sum of each column
4 Exercise
- We will evaluate different summarization methods (Maxquant maxLFQ, median and robust model based) in the tutorial session before discussing on their advantages/disadvantages.
- Can you anticipate on potential problems related to the summarization?
5 Software & code
Our R/Bioconductor package msqrob2 can be used in R markdown scripts or with a GUI/shinyApp in the msqrob2gui package.
The GUI is intended as a introduction to the key concepts of proteomics data analysis for users who have no experience in R.
However, learning how to code data analyses in R markdown scripts is key for open en reproducible science and for reporting your proteomics data analyses and interpretation in a reproducible way.
More information on our tools can be found in our papers (L. J. Goeminne, Gevaert, and Clement 2016), (L. J. E. Goeminne et al. 2020) and (Sticker et al. 2020). Please refer to our work when using our tools.
5.1 Code
Data infrastructure
Import proteomics data
Preprocessing
Log-transformation
Filtering
Normalisation
Summarization
