Technical details on linear regression for proteomics when starting from summarized protein expression values
Lieven Clement
statOmics, Ghent University (https://statomics.github.io)
1 Preamble
Read stored results for heart data
library(tidyverse)
library(limma)
library(QFeatures)
library(msqrob2)
pe <- readRDS(
url(
"https://raw.githubusercontent.com/statOmics/SGA2020/gh-pages/assets/peHeart.rds",
"rb")
)2 Linear regression
- Consider a vector of predictors \(\mathbf{x}_i=(x_1,\ldots,x_{p-1})\) and
- a real-valued response \(Y_i\)
- with \(i = 1, \ldots, n\)
- then the linear regression model can be written as \[ Y_i=f(\mathbf{x}) +\epsilon=\beta_0+\sum\limits_{j=1}^{p-1} x_{ij}\beta + \epsilon_i \] with i.i.d. \(\epsilon_i\sim N(0,\sigma^2)\)
- \(n\) observations \((\mathbf{x}_1,y_1) \ldots (\mathbf{x}_n,y_n)\)
- Regression in matrix notation \[\mathbf{Y}=\mathbf{X\beta} + \boldsymbol{\epsilon}\] with \(\mathbf{Y}=\left[\begin{array}{c}y_1\\ \vdots\\y_n\end{array}\right]\), \(\mathbf{X}=\left[\begin{array}{cccc} 1&x_{11}&\ldots&x_{1p-1}\\ \vdots&\vdots&&\vdots\\ 1&x_{n1}&\ldots&x_{np-1} \end{array}\right]\), \(\boldsymbol{\beta}=\left[\begin{array}{c}\beta_0\\ \vdots\\ \beta_{p-1}\end{array}\right]\) and \(\boldsymbol{\epsilon}=\left[\begin{array}{c} \epsilon_1 \\ \vdots \\ \epsilon_n\end{array}\right]\)
2.1 Least Squares (LS)
Minimize the residual sum of squares \[\begin{eqnarray*} RSS(\boldsymbol{\beta})&=&\sum\limits_{i=1}^n e^2_i\\ &=&\sum\limits_{i=1}^n \left(y_i-\beta_0-\sum\limits_{j=1}^p x_{ij}\beta_j\right)^2 \end{eqnarray*}\]
or in matrix notation \[\begin{eqnarray*} RSS(\boldsymbol{\beta})&=&(\mathbf{Y}-\mathbf{X\beta})^T(\mathbf{Y}-\mathbf{X\beta})\\ &=&\Vert \mathbf{Y}-\mathbf{X\beta}\Vert^2 \end{eqnarray*}\] with the \(L_2\)-norm of a \(p\)-dim. vector \(v\) \(\Vert \mathbf{v} \Vert=\sqrt{v_1^2+\ldots+v_p^2}\)
\(\rightarrow\) \(\hat{\boldsymbol{\beta}}=\text{argmin}_\beta \Vert \mathbf{Y}-\mathbf{X\beta}\Vert^2\)}
2.1.1 Minimize RSS
\[ \begin{array}{ccc} \frac{\partial RSS}{\partial \boldsymbol{\beta}}&=&\mathbf{0}\\\\ \frac{(\mathbf{Y}-\mathbf{X\beta})^T(\mathbf{Y}-\mathbf{X\beta})}{\partial \boldsymbol{\beta}}&=&\mathbf{0}\\\\ -2\mathbf{X}^T(\mathbf{Y}-\mathbf{X\beta})&=&\mathbf{0}\\\\ \mathbf{X}^T\mathbf{X\beta}&=&\mathbf{X}^T\mathbf{Y}\\\\ \hat{\boldsymbol{\beta}}&=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\mathbf{Y} \end{array} \]
2.1.1.1 Heart example
y <- assay(pe[["proteinRobust"]])[2,]
fit <- lm(y ~ location*tissue + patient, data = colData(pe), x = TRUE)
head(fit$x,4)## (Intercept) locationR tissueV patient4 patient8 locationR:tissueV
## Intensity.LA3 1 0 0 0 0 0
## Intensity.LA4 1 0 0 1 0 0
## Intensity.LA8 1 0 0 0 1 0
## Intensity.LV3 1 0 1 0 0 0The model matrix can also be obtained without fitting the model:
X <- model.matrix(~ location * tissue + patient, colData(pe))
head(X,4)## (Intercept) locationR tissueV patient4 patient8 locationR:tissueV
## Intensity.LA3 1 0 0 0 0 0
## Intensity.LA4 1 0 0 1 0 0
## Intensity.LA8 1 0 0 0 1 0
## Intensity.LV3 1 0 1 0 0 0Least squares:
betas <- solve(t(X)%*%X) %*% t(X) %*% y
cbind(fit$coef, betas)## [,1] [,2]
## (Intercept) 26.57559360 26.57559360
## locationR 0.20636497 0.20636497
## tissueV 0.12756675 0.12756675
## patient4 0.30592435 0.30592435
## patient8 -0.33432636 -0.33432636
## locationR:tissueV -0.04130843 -0.041308432.1.2 Variance Estimator?
\[ \begin{array}{ccl} \hat{\boldsymbol{\Sigma}}_{\hat{\boldsymbol{\beta}}} &=&\text{var}\left[(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\mathbf{Y}\right]\\\\ &=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\text{var}\left[\mathbf{Y}\right]\mathbf{X}(\mathbf{X}^T\mathbf{X})^{-1}\\\\ &=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T(\mathbf{I}\sigma^2)\mathbf{X}(\mathbf{X}^T\mathbf{X})^{-1} \\\\ &=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\mathbf{I}\quad\mathbf{X}(\mathbf{X}^T\mathbf{X})^{-1}\sigma^2\\\\ %\hat{\boldmath{\Sigma}}_{\hat{\boldsymbol{\beta}}}&=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\var\left[\mathbf{Y}\right](\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}\\ &=&(\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}^T\mathbf{X}(\mathbf{X}^T\mathbf{X})^{-1}\sigma^2\\\\ &=&(\mathbf{X}^T\mathbf{X})^{-1}\sigma^2 \end{array} \]
2.1.2.1 Heart example
summary(fit)$cov.unscaled * sigma(fit)^2## (Intercept) locationR tissueV patient4
## (Intercept) 0.3938774 -2.625849e-01 -2.625849e-01 -1.969387e-01
## locationR -0.2625849 5.251699e-01 2.625849e-01 1.549906e-16
## tissueV -0.2625849 2.625849e-01 5.251699e-01 1.549906e-16
## patient4 -0.1969387 1.549906e-16 1.549906e-16 3.938774e-01
## patient8 -0.1969387 1.249600e-16 1.249600e-16 1.969387e-01
## locationR:tissueV 0.2625849 -5.251699e-01 -5.251699e-01 -1.332518e-16
## patient8 locationR:tissueV
## (Intercept) -1.969387e-01 2.625849e-01
## locationR 1.249600e-16 -5.251699e-01
## tissueV 1.249600e-16 -5.251699e-01
## patient4 1.969387e-01 -1.332518e-16
## patient8 3.938774e-01 -1.236848e-16
## locationR:tissueV -1.236848e-16 1.050340e+00n <- nrow(X)
p <- ncol(X)
mse <- sum((y-X%*%betas)^2)/(n-p)
SigmaBeta <- solve(t(X)%*%X) * mse
SigmaBeta## (Intercept) locationR tissueV patient4 patient8
## (Intercept) 0.3938774 -0.2625849 -0.2625849 -0.1969387 -0.1969387
## locationR -0.2625849 0.5251699 0.2625849 0.0000000 0.0000000
## tissueV -0.2625849 0.2625849 0.5251699 0.0000000 0.0000000
## patient4 -0.1969387 0.0000000 0.0000000 0.3938774 0.1969387
## patient8 -0.1969387 0.0000000 0.0000000 0.1969387 0.3938774
## locationR:tissueV 0.2625849 -0.5251699 -0.5251699 0.0000000 0.0000000
## locationR:tissueV
## (Intercept) 0.2625849
## locationR -0.5251699
## tissueV -0.5251699
## patient4 0.0000000
## patient8 0.0000000
## locationR:tissueV 1.0503398range(SigmaBeta - summary(fit)$cov.unscaled * sigma(fit)^2)## [1] -3.996803e-15 7.993606e-15data.frame(summary(fit)$coef[,1:2], betas = betas, seBetas = diag(SigmaBeta)^.5)2.2 Contrasts
When we assess a contrast we assess a linear combination of model parameters:
\[ H_0: \mathbf{L^T\beta} = 0 \text{ vs } H_1: \mathbf{L^T\beta} \neq 0 \]
Estimator of Contrast?
\[\mathbf{L}^T\hat{\boldsymbol{\beta}}\]
Variance?
\[ \boldsymbol{\Sigma}_{\mathbf{L}\hat{\boldsymbol{\beta}}}=\mathbf{L}^T\boldsymbol{\Sigma}_{\hat{\boldsymbol{\beta}}}\mathbf{L} \]
2.2.1 Heart example
L <- makeContrast(
c(
"tissueV = 0",
"tissueV + locationR:tissueV = 0",
"tissueV + 0.5*locationR:tissueV = 0","locationR:tissueV = 0"),
parameterNames =
rowData(pe[["proteinRobust"]])$msqrobModels[[2]] %>%
getCoef %>%
names
)
L## tissueV tissueV + locationR:tissueV
## (Intercept) 0 0
## locationR 0 0
## tissueV 1 1
## patient4 0 0
## patient8 0 0
## locationR:tissueV 0 1
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## (Intercept) 0.0 0
## locationR 0.0 0
## tissueV 1.0 0
## patient4 0.0 0
## patient8 0.0 0
## locationR:tissueV 0.5 1contrasts <- t(L) %*% betas
SigmaContrasts <- t(L) %*% SigmaBeta %*% L
seContrasts <- SigmaContrasts %>%
diag %>%
sqrtComparison with lm and glht results
library(multcomp)## Loading required package: mvtnorm## Loading required package: survival## Loading required package: TH.data## Loading required package: MASS##
## Attaching package: 'MASS'## The following object is masked from 'package:dplyr':
##
## select##
## Attaching package: 'TH.data'## The following object is masked from 'package:MASS':
##
## geyserfitGlht <- glht(fit, linfct = t(L))
summary(fitGlht, test = adjusted("none"))##
## Simultaneous Tests for General Linear Hypotheses
##
## Fit: lm(formula = y ~ location * tissue + patient, data = colData(pe),
## x = TRUE)
##
## Linear Hypotheses:
## Estimate Std. Error t value Pr(>|t|)
## tissueV == 0 0.12757 0.72469 0.176 0.866
## tissueV + locationR:tissueV == 0 0.08626 0.72469 0.119 0.909
## tissueV + 0.5 * locationR:tissueV == 0 0.10691 0.51243 0.209 0.842
## locationR:tissueV == 0 -0.04131 1.02486 -0.040 0.969
## (Adjusted p values reported -- none method)data.frame(contrasts, seContrasts)Note, that the power for assessing \(\log_2\) FC between ventriculum and atrium left and right is the same. Indeed, the standard errors are equal for both effects.
Note, that the power for assessing \(\log_2\) FC between ventriculum and atrium over both heart regions is higher than when assessing the effect left or right.
- Indeed, the standard error is a factor \(\sqrt{2}\) smaller for the former effect
- We intuitively can explain this because we can use all samples (double the number of samples) to assess the average effect.
- Hence the variance is a factor two smaller, and the se with a factor \(\sqrt{2}\)
Note, that we have the lowest power to pick up an interaction effect. Indeed, the se is a factor \(\sqrt{2}\) larger than for the ventriculum - atrium effect left or right and a factor 2 larger than for the average effect between ventriculum and atrium.
seContrasts / seContrasts[1]## tissueV tissueV + locationR:tissueV
## 1.0000000 1.0000000
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.7071068 1.4142136sqrt(2)## [1] 1.4142141/sqrt(2)## [1] 0.70710682.2.2 t-tests
When the assumptions of the linear model hold \[ \hat{\boldsymbol{\beta}} \sim MVN\left[\boldsymbol{\beta},\left(\mathbf{X}^T\mathbf{X}\right)^{-1}\sigma^2\right] \]
Hence, \[ \mathbf{L}^T\hat{\boldsymbol{\beta}} \sim MVN\left[\mathbf{L}^T\boldsymbol{\beta},\mathbf{L}^T\left[\left(\mathbf{X}^T\mathbf{X}\right)^{-1}\sigma^2\right]\mathbf{L}\right] \]
We estimate \(\sigma^2\) by MSE \[\hat{\sigma}^2=\frac{\mathbf{e}^T\mathbf{e}}{n-p} \rightarrow \hat{\boldsymbol{\Sigma}}_{\hat{\boldsymbol{\beta}}}=\left(\mathbf{X}^T\mathbf{X}\right)^{-1}\hat\sigma^2\]
When we test one contrast at the time (e.g. the \(k^\text{th}\) contrast) the statistic reduces to
\[T=\frac{\mathbf{L}_k^T\hat{\boldsymbol{\beta}}}{\sqrt{\left(\mathbf{L}^T_k\hat{\boldsymbol{\Sigma}}_{\hat{\boldsymbol{\beta}}}\mathbf{L}_k\right)}} \underset{H_0}{\sim} t_{n-p}\] follows a t distribution with n-p degrees of freedom under \(H_0: \mathbf{L}^T_k\hat{\boldsymbol{\beta}}=0\)
2.2.2.1 Heart example
tContrasts <- contrasts/seContrasts
pContrasts <- pt(abs(tContrasts),
df = n - p,
lower.tail = FALSE) * 2Comparison with lm and glht results
summary(fitGlht, test = adjusted("none"))##
## Simultaneous Tests for General Linear Hypotheses
##
## Fit: lm(formula = y ~ location * tissue + patient, data = colData(pe),
## x = TRUE)
##
## Linear Hypotheses:
## Estimate Std. Error t value Pr(>|t|)
## tissueV == 0 0.12757 0.72469 0.176 0.866
## tissueV + locationR:tissueV == 0 0.08626 0.72469 0.119 0.909
## tissueV + 0.5 * locationR:tissueV == 0 0.10691 0.51243 0.209 0.842
## locationR:tissueV == 0 -0.04131 1.02486 -0.040 0.969
## (Adjusted p values reported -- none method)data.frame(contrasts, seContrasts, tContrasts, pContrasts)3 Robust regression
With msqrob2 we perform robust regression to estimate the model parameters of the regression model
No normality assumption needed
Robust fit minimises the maximal bias of the estimators
CI and statistical tests are based on asymptotic theory
If \(\epsilon\) is normal, the M-estimators have a high efficiency!
ordinary least squares (OLS): minimize loss function \[\sum\limits_{i=1}^n (y_i-\mathbf{x}_i^T\boldsymbol{\beta})^2\]
M-estimation: minimize loss function \[\sum\limits_{i=1}^n \rho\left(y_i-\mathbf{x}_i^T\boldsymbol{\beta}\right)\] with
- \(\rho\) is symmetric, i.e. \(\rho(z)=\rho(-z)\)
- \(\rho\) has a minimum at \(\rho(0)=0\), is positive for all \(z\neq 0\)
- \(\rho(z)\) increases as \(\vert z\vert\) increases
The estimator \(\hat{\mu}\) is also the solution to the equation \[ \sum_{i=1}^n \Psi(y_i - \mathbf{x}_i\boldsymbol{\beta}) =0, \] where \(\Psi\) is the derivative of \(\rho\). For \(\hat{\beta}\) possessing the robustness property, \(\Psi\) should be bounded.
Example: least squares
\(\rho(z) = z^2\), and thus \(\Psi(z)=2z\) (unbounded!). Not robust!
\(\hat{\boldsymbol{\beta}}\) is the solution of \[ \sum_{i=1}^n 2 \mathbf{x}_i (y_i - \mathbf{x}_i^T\boldsymbol{\beta}) = 0 \text{ or } \hat{\boldsymbol{\beta}} = (\mathbf{X}^T\mathbf{X})^{-1}\mathbf{X}\mathbf{y} \] with \(\mathbf{X}=[\mathbf{x}_1 \ldots \mathbf{x}_G]^T\)
When a location and a scale parameter, say \(\sigma\), have to be estimated simultaneously, we write \[ (\hat{\boldsymbol{\beta}},\hat{\sigma}) = \text{ArgMin}_{\boldsymbol{\beta},\sigma} \sum_{i=1}^n \rho\left(\frac{y_i - \mathbf{x}_i^T\boldsymbol{\beta}}{\sigma}\right) \text{ and } \sum_{i=1}^n \Psi\left(\frac{y_i - \mathbf{x}_i^T\boldsymbol{\beta}}{\sigma}\right) =0. \]
Define \(u_i = \frac{y_i - \mathbf{x}_i^T\boldsymbol{\beta}}{\sigma}\). The last estimation equation is equivalent to \[ \sum_{i=1}^n w(u_i) u_i = 0 , \] with weight function \(w(u)=\Psi(u)/u\). This is the typical form that appears when solving the iteratively reweighted least squares problem, \[ (\hat{\boldsymbol{\beta}},\hat{\sigma}) = \text{ArgMin}_{\mu,\sigma} \sum_{i=1}^n w(u_i^{(k-1)}) \left(u_i^{(k)}\right)^2 , \] where \(k\) represents the iteration number.
3.1 Some Examples of Robust Functions
PhD thesis Bolstad 2004
3.2 The \(\rho\) functions
PhD thesis Bolstad 2004
3.2.1 Common \(\Psi\)-Functions
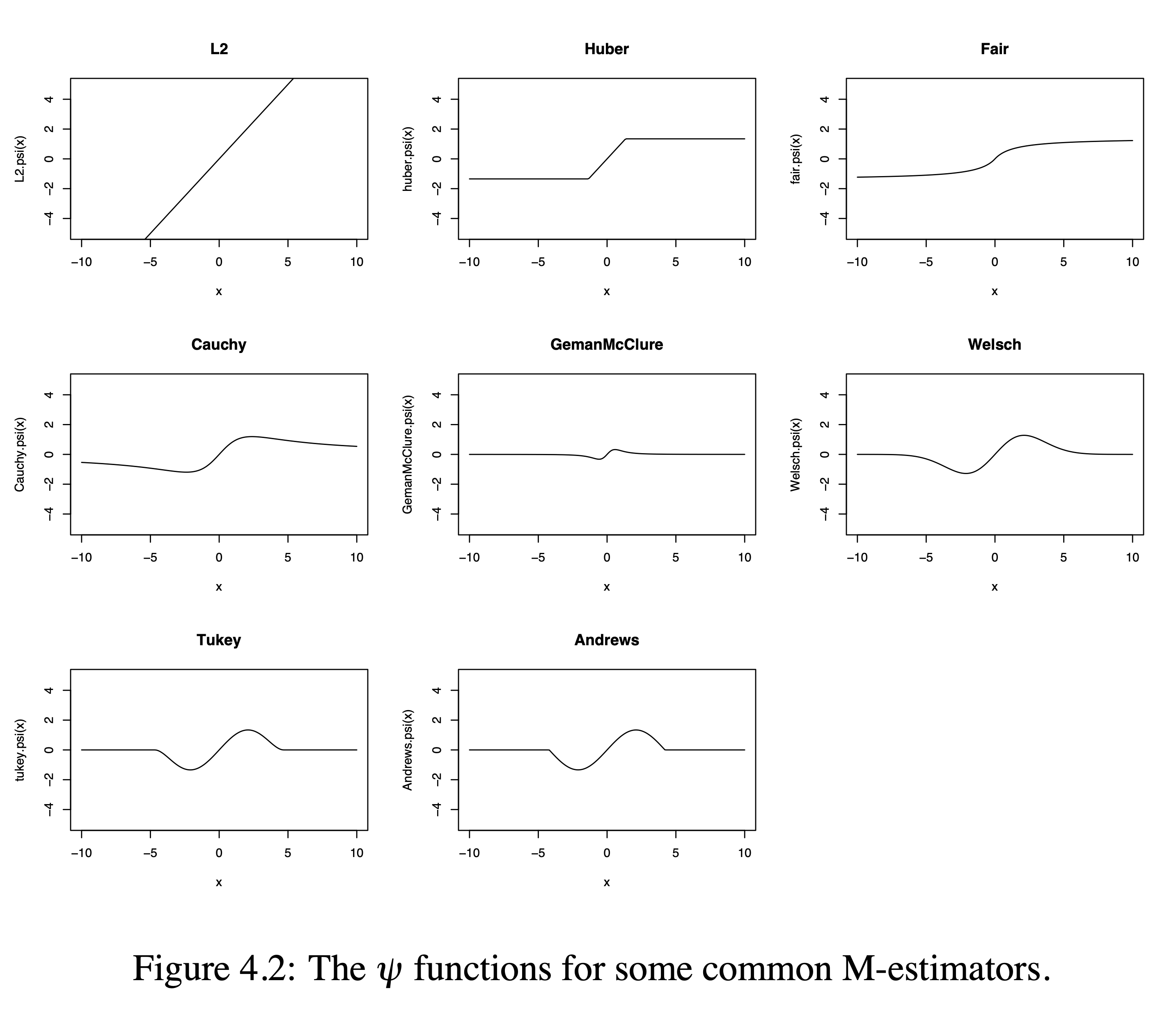
PhD thesis Bolstad 2004
3.2.2 Corresponding Weight Functions
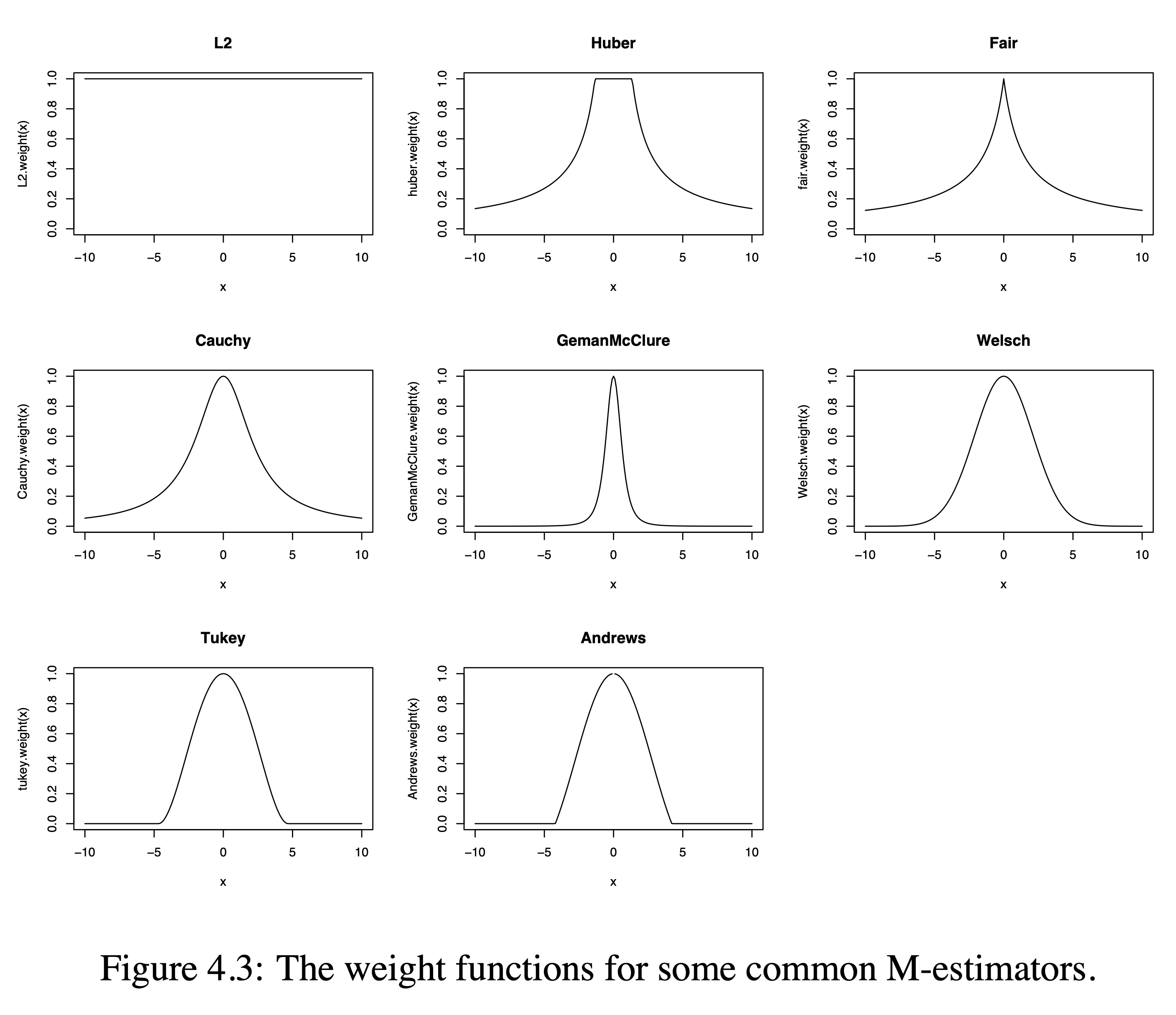
PhD thesis Bolstad 2004
library("MASS")
rfit <- rlm(y ~ location * tissue + patient, colData(pe), maxit=1)## Warning in rlm.default(x, y, weights, method = method, wt.method = wt.method, :
## 'rlm' failed to converge in 1 stepsqplot(fit$coefficient[-1],
rfit$coefficient[-1],
xlab="fit",
ylab="robust fit") +
geom_abline() +
xlim(range(c(fit$coefficient[-1],rfit$coefficient[-1]))) +
ylim(range(c(fit$coefficient[-1],rfit$coefficient[-1])))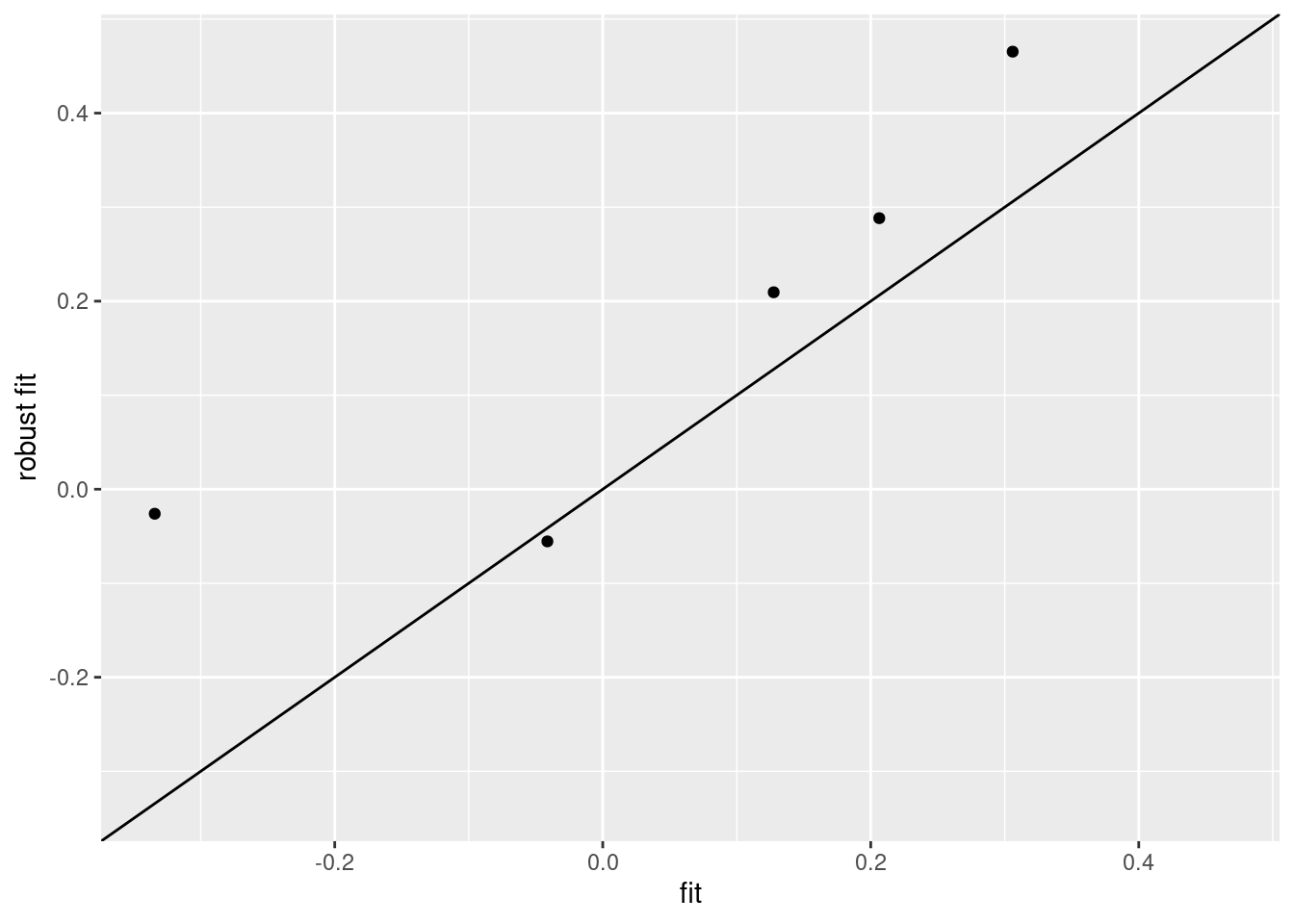
rfit$w## [1] 0.9516397 1.0000000 0.6628477 1.0000000 1.0000000 1.0000000 1.0000000
## [8] 1.0000000 1.0000000 0.4805187 1.0000000 0.4344049plot(
rfit$fitted,
rfit$res,
cex=rfit$w,
pch=19,col=2,
cex.lab=1.5,
cex.axis=1.5,
ylab="residuals",
xlab="fit")
points(rfit$fitted, rfit$res , cex= 1.5)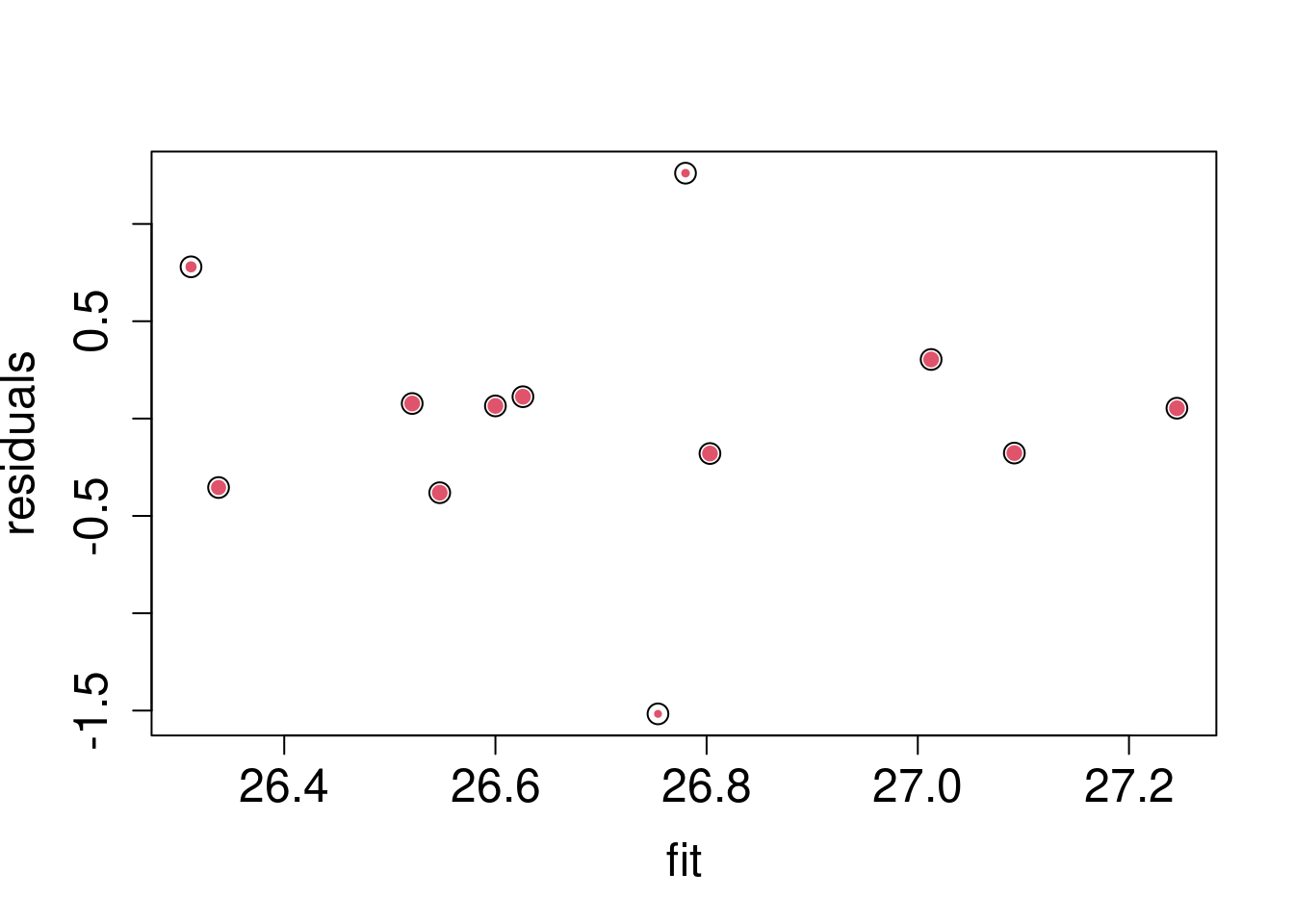
summary(fit)##
## Call:
## lm(formula = y ~ location * tissue + patient, data = colData(pe),
## x = TRUE)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.29698 -0.32767 0.04041 0.24905 1.17251
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 26.57559 0.62760 42.345 1.16e-08 ***
## locationR 0.20636 0.72469 0.285 0.785
## tissueV 0.12757 0.72469 0.176 0.866
## patient4 0.30592 0.62760 0.487 0.643
## patient8 -0.33433 0.62760 -0.533 0.613
## locationR:tissueV -0.04131 1.02486 -0.040 0.969
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.8876 on 6 degrees of freedom
## Multiple R-squared: 0.1687, Adjusted R-squared: -0.524
## F-statistic: 0.2436 on 5 and 6 DF, p-value: 0.9286summary(rfit)##
## Call: rlm(formula = y ~ location * tissue + patient, data = colData(pe),
## maxit = 1)
## Residuals:
## Min 1Q Median 3Q Max
## -1.51703 -0.22326 0.05909 0.16009 1.26070
##
## Coefficients:
## Value Std. Error t value
## (Intercept) 26.3378 0.5391 48.8585
## locationR 0.2883 0.6225 0.4631
## tissueV 0.2095 0.6225 0.3365
## patient4 0.4654 0.5391 0.8633
## patient8 -0.0261 0.5391 -0.0484
## locationR:tissueV -0.0555 0.8803 -0.0630
##
## Residual standard error: 0.4189 on 6 degrees of freedomrowData(pe[["proteinRobust"]])$msqrobModels[[2]] %>% getCoef## (Intercept) locationR tissueV patient4
## 26.33779939 0.28826213 0.20946392 0.46536955
## patient8 locationR:tissueV
## -0.02608046 -0.055501113.3 Understanding implementation of robust regression
3.3.1 Simulate 20 observations from a linear model with errors that follow a normal distribution
set.seed <- 112358
nobs <- 20
sdy <- 1
xsim <- seq(0, 1, length.out = nobs)
ysim <- 10 + 5*xsim + rnorm(nobs, sd = sdy)3.3.2 add outlier at high leverage point
ysim[nobs] <- 73.3.3 fit linear model
ols <- lm(ysim ~ xsim)3.3.4 fit robust linear model
library(MASS)
mEst <- rlm(ysim ~ xsim)3.3.4.1 plot results
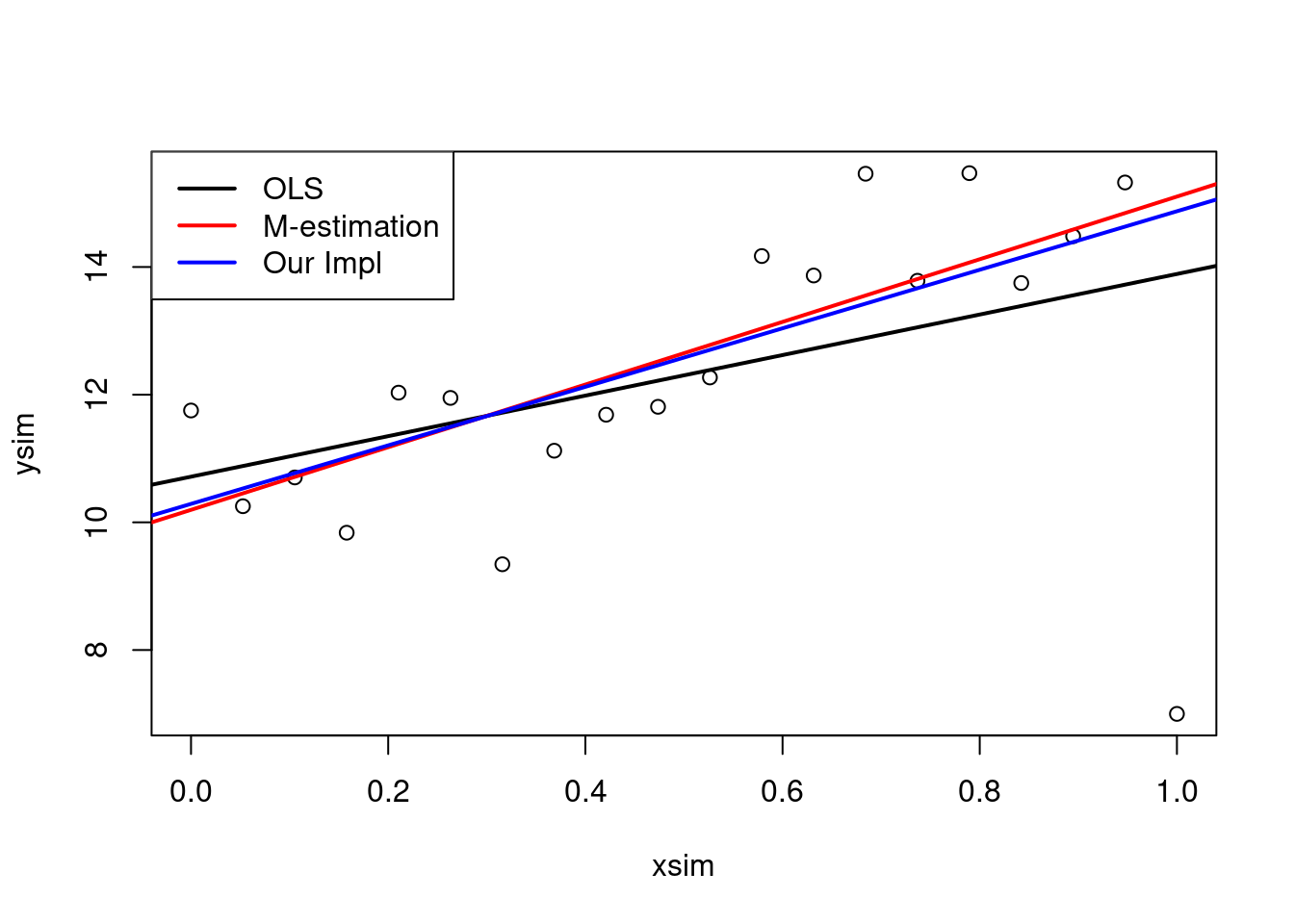
plot(xsim, ysim)
abline(ols, lwd = 2)
abline(mEst, col = "red", lwd = 2)
legend("topleft",
legend = c("OLS", "M-estimation"),
lwd = 2,
col = 1:2)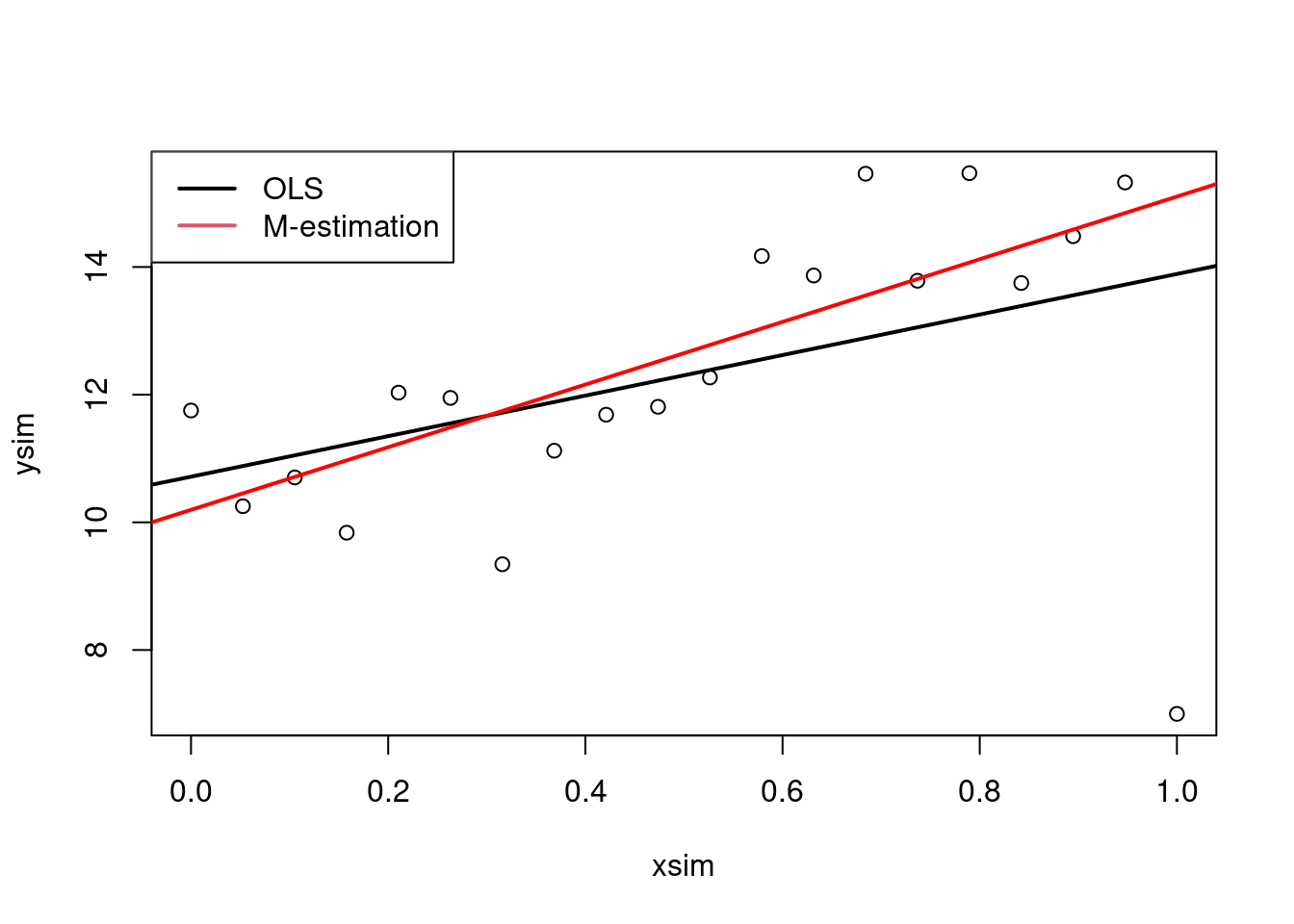
round(mEst$w,3)## [1] 0.826 1.000 1.000 1.000 1.000 1.000 0.535 1.000 1.000 1.000 1.000 1.000
## [13] 1.000 0.674 1.000 0.919 1.000 1.000 1.000 0.159The plot clearly shows that the outlier has a high impact on the slope estimate. This is because the outlier is at a high leverage point, i.e. far from the average covariate pattern.
3.3.5 Implement it yourself
3.3.5.1 start from ols fit
lmMod <- ols3.3.5.2 Use robust variance estimator to calculate the z
res <- lmMod$res
stdev <- mad(res)
stdev## [1] 1.153029median(abs(res-median(res)))*1.4826## [1] 1.153029z <- res/stdev3.3.5.3 Calculate weights use psi.huber function
w <- psi.huber(z)
plot(xsim, ysim)
plot(xsim, lmMod$res, cex = w, pch = 19, col = "red")
points(xsim,lmMod$res, cex = 1.5)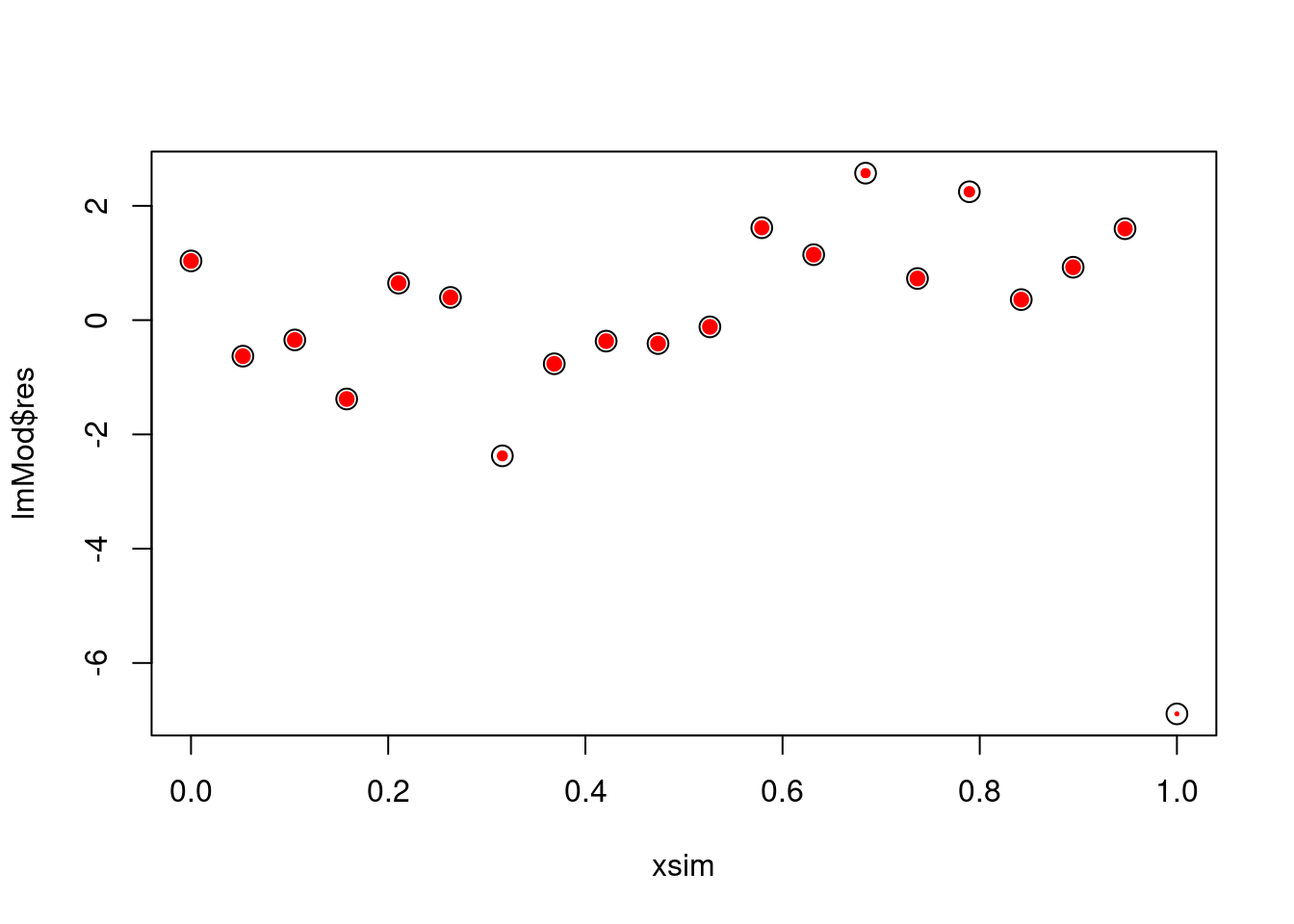
3.3.5.4 Perform a weighted regression use lm with weights=w
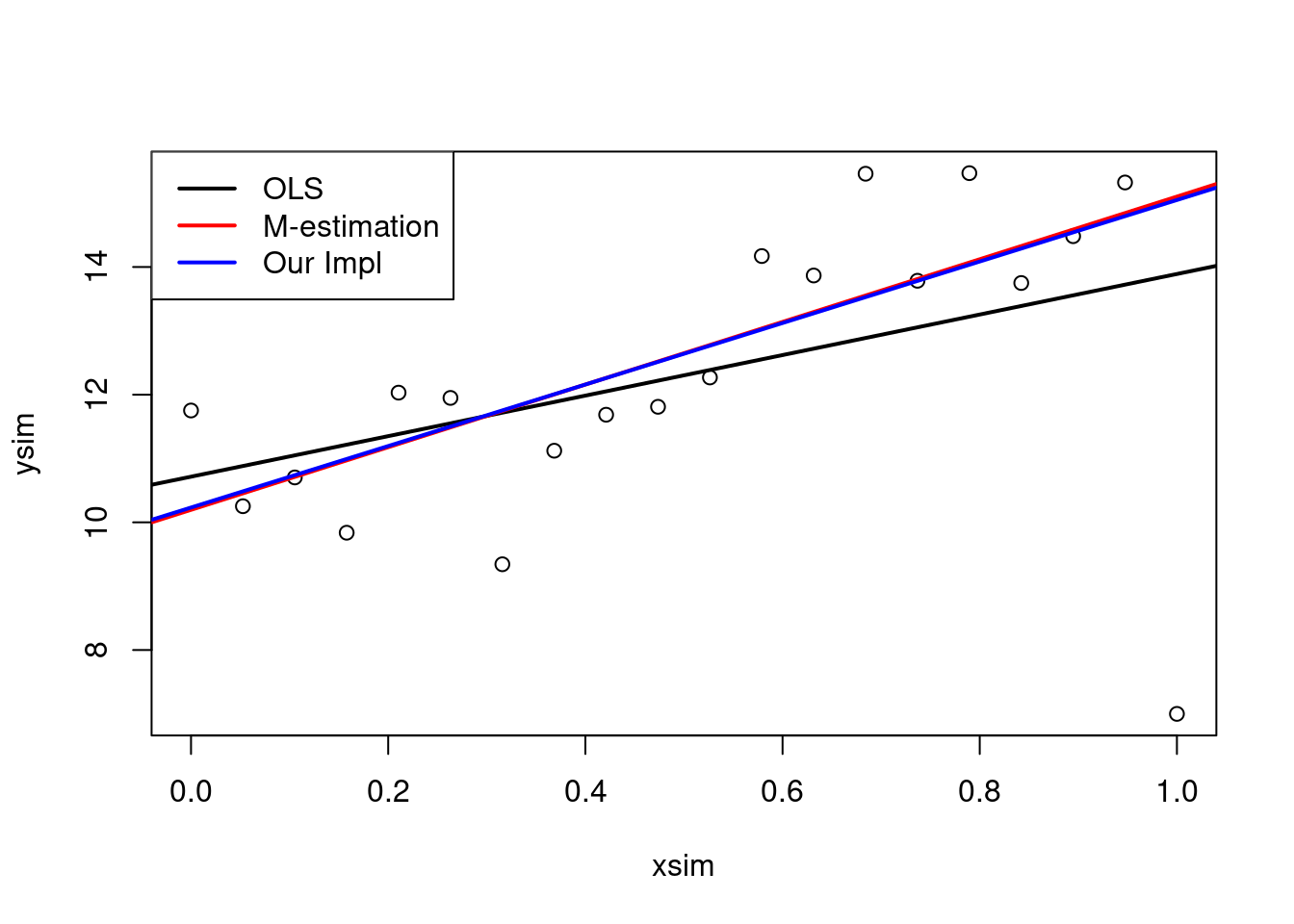
lmMod <- lm(ysim~xsim, weights = w)3.3.5.5 Plot results
plot(xsim, ysim)
abline(ols, lwd = 2)
abline(mEst, col = "red", lwd = 2)
abline(lmMod, col = "blue", lwd = 2)
legend("topleft",
legend = c("OLS","M-estimation","Our Impl"),
lwd = 2,
col = c("black", "red", "blue"))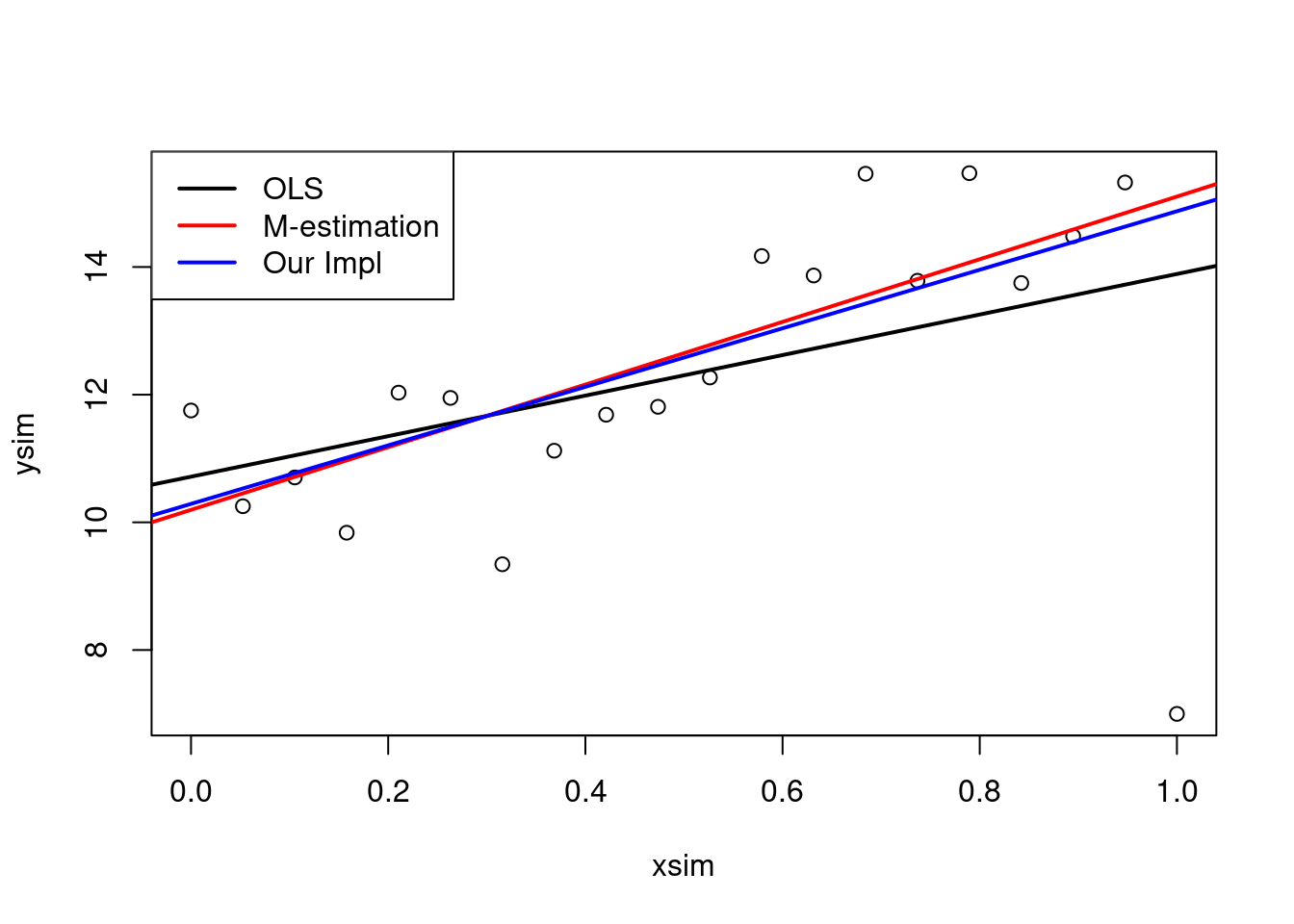
3.3.5.6 Repeat this many times
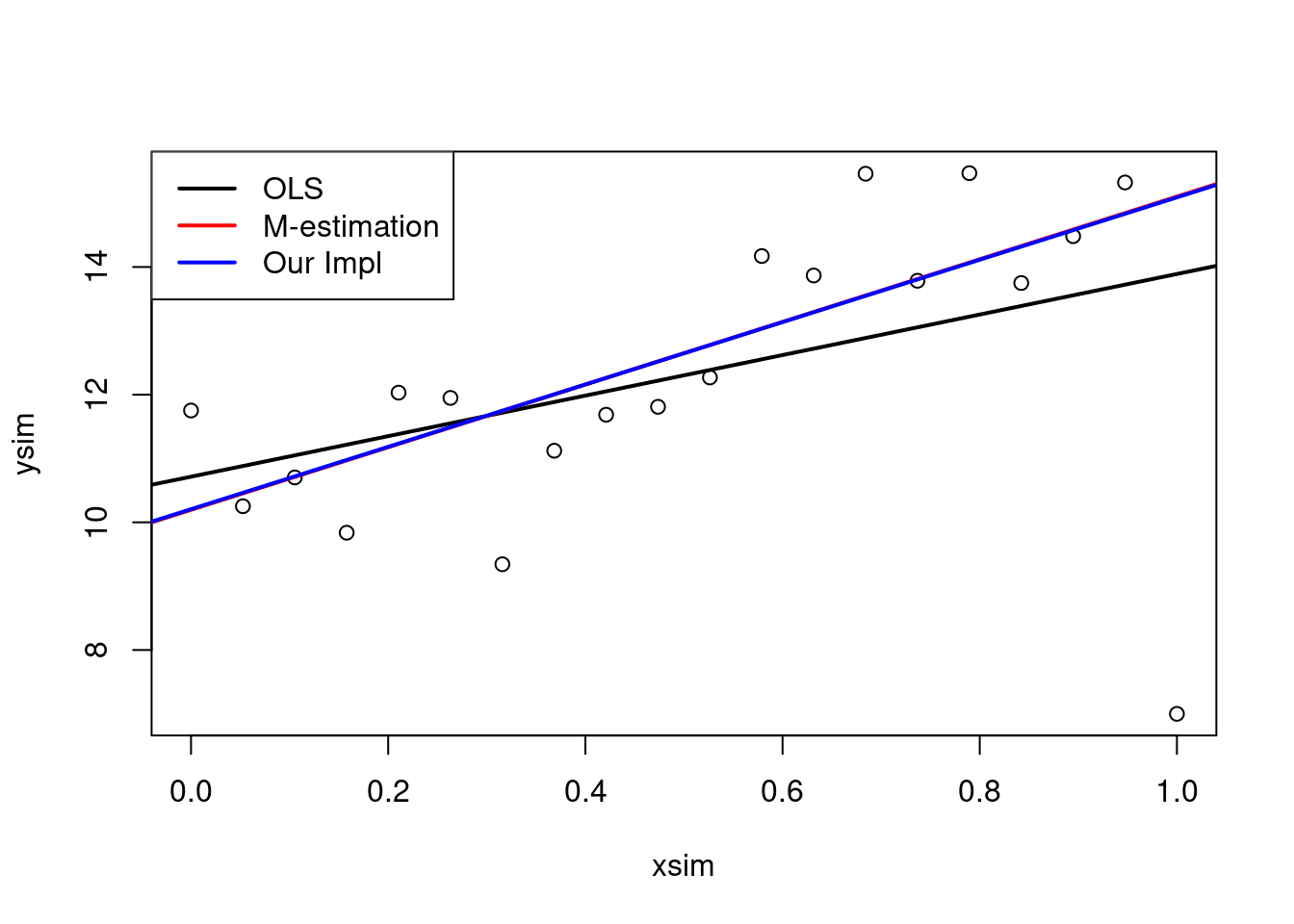
lmMod <- ols
for (k in 1:3)
{
######repeat this part several times until convergence
#use robust variance estimator to calculate the z
res <- lmMod$res
stdev <- mad(res)
median(abs(res-median(res)))*1.4826
z <- res/stdev
#calculate weights
#use psi.huber function
w <- psi.huber(z)
#perform a weighted regression use lm with weights=w
lmMod <- lm(ysim ~ xsim, weights = w)
#plot results
plot(xsim,ysim)
abline(ols, lwd = 2)
abline(mEst, col = "red", lwd = 2)
abline(lmMod, col = "blue", lwd = 2)
legend("topleft",
legend = c("OLS","M-estimation","Our Impl"),
lwd = 2,
col = c("black", "red", "blue")
)
####################################
}
4 Empirical Bayes/Moderated \(t\)-test.
A general class of moderated test statistics is given by
\[\tilde{T}_p = \frac{\mathbf{L}_k \hat{ \boldsymbol{\beta_p}}}{\mathbf{L}_k^T(\mathbf{X}^T\mathbf{WX})^{-1}\mathbf{L}_k^T \tilde{s}_p^2}\]
where \(\tilde{s}_p\) is a moderated variance estimator.
Simple approach: set \(\tilde{s}_p=s_p + s_0\): simply add a small positive constant to the denominator of the t-statistic
theory provides formal framework for borrowing strength across genes or proteins, e.g. popular bioconductor package \[\tilde{s}_g=\sqrt{\frac{d_ps_p^2+d_0s_0^2}{d_g+d_0}},\] and the moderated t-statistic is t-distributed with \(d_0+d_g\) degrees of freedom under the null hypothesis \(H_0: \mathbf{L}\boldsymbol{\beta}=0\).
- Note, that the degrees of freedom increase by borrowing strength across proteins.
4.1 Intermezzo: Bayesian Methods
Frequentists consider data as random and population parameters as fixed but unknown
In Bayesian viewpoint a person has prior beliefs about the population parameters and the uncertainty on this prior beliefs are represented by a probability distribution placed on this parameter.
- This distribution reflects the person’s subjective prior opinion about plausible values of the parameter.
- And is referred to as the prior \(g(\boldsymbol{\theta})\).
Bayesian thinking will update the prior information on the population parameters by confronting the model to data (\(\mathbf{Y}\)).
By using Bayes Theorem this results in a posterior distribution on the model parameters.
\[ g(\boldsymbol{\theta}\vert\mathbf{Y})=\frac{f(Y\vert \boldsymbol{\theta})g(\boldsymbol{\theta})}{\int f(Y\vert \boldsymbol{\theta}) g(\boldsymbol{\theta}) d\boldsymbol{\theta}} \text{ }\left(\text{ posterior}=\frac{\text{prior} \times \text{ likelihood}}{\text{Marginal distribution}}\right) \]
4.2 Limma approach
Developed for gene expression analysis with micro arrays. Let g be the index for gene g. \[ \begin{array}{cc} &\beta_{gk}\vert \sigma^2_g,\beta_{gk}\neq 0 \sim N(0,v_{0k}\sigma_g^2)\\\\ \text{Prior}\\ &\frac{1}{\sigma^2_g}\sim s^2_0\frac{\chi^2_{d_0}}{d_0}\\\\\\\\ &\hat \beta_{gk} | \beta_{gk} , \sigma_g^2 \sim N( \beta_{gk} , v_{gk}\sigma_g^2)\\\\ \text{Data}\\ &s_g^2\sim \sigma^2_g\frac{\chi^2_{d_g}}{d_g}\\\\ \end{array} \]
4.3 Limma approach
Under this assumption, it can be shown that
Posterior Mean for the variance parameter: \[\tilde{s}^2_p = \text{E}\left[\sigma^2_p\vert s_p^2\right]=\frac{d_0 s_0^2+d_ps_p^2}{d_0+d_p}\]
\[\tilde{T}_p=\frac{\mathbf{L}_k \hat{ \boldsymbol{\beta_p}}}{\mathbf{L}_k^T(\mathbf{X}^T\mathbf{WX})^{-1}\mathbf{L}_k^T \tilde{s}_p^2}\]
is t-distributed under \(H_0: \mathbf{L}_j\boldsymbol{\beta} = 0\)
\[\tilde{T}_p \vert H_0 \sim t(d_0 + d_p)\]
4.4 Empirical Bayes
- A fully Bayesian
- would define the prior distribution by carefully choosing the prior parameters based on prior knowledge on the process
- would confront the prior to the data and performs inference using the posterior distribution of the model parameters.
- In an empirical Bayesian approach one estimates the prior parameters based on the data.
- In Limma moment estimators for \(s_0\) and \(d_0\) are derived using the information on the gene (protein) wise variances of all genes (proteins).
- In Limma one also does not work with the full posterior distribution for the variances, but with the maximum a-posterior estimate.
4.5 Illustration
We borrow strength across proteins by
- placing a scaled \(\chi^2\) prior: \(\chi^2(s_o,d_0)\) on the precisions (\(1/\sigma^2_p\))
- estimating the prior parameters \(s_0\) and \(df_0\)
- replacing the estimated protein-wise variances (\(s_p^2\)) with the maximum a-posteriori variance \[\tilde{s}_p = \frac{d_p s^2_p + d_0 s_0^2}{d_p+d_0}\]
sd <- sapply(
rowData(pe[["proteinRobust"]])$msqrobModels,
getSigma) %>%
na.exclude
sdPost <- sapply(
rowData(pe[["proteinRobust"]])$msqrobModels,
getSigmaPosterior) %>%
na.exclude
p1 <- qplot(sd,sdPost) +
geom_abline()
p1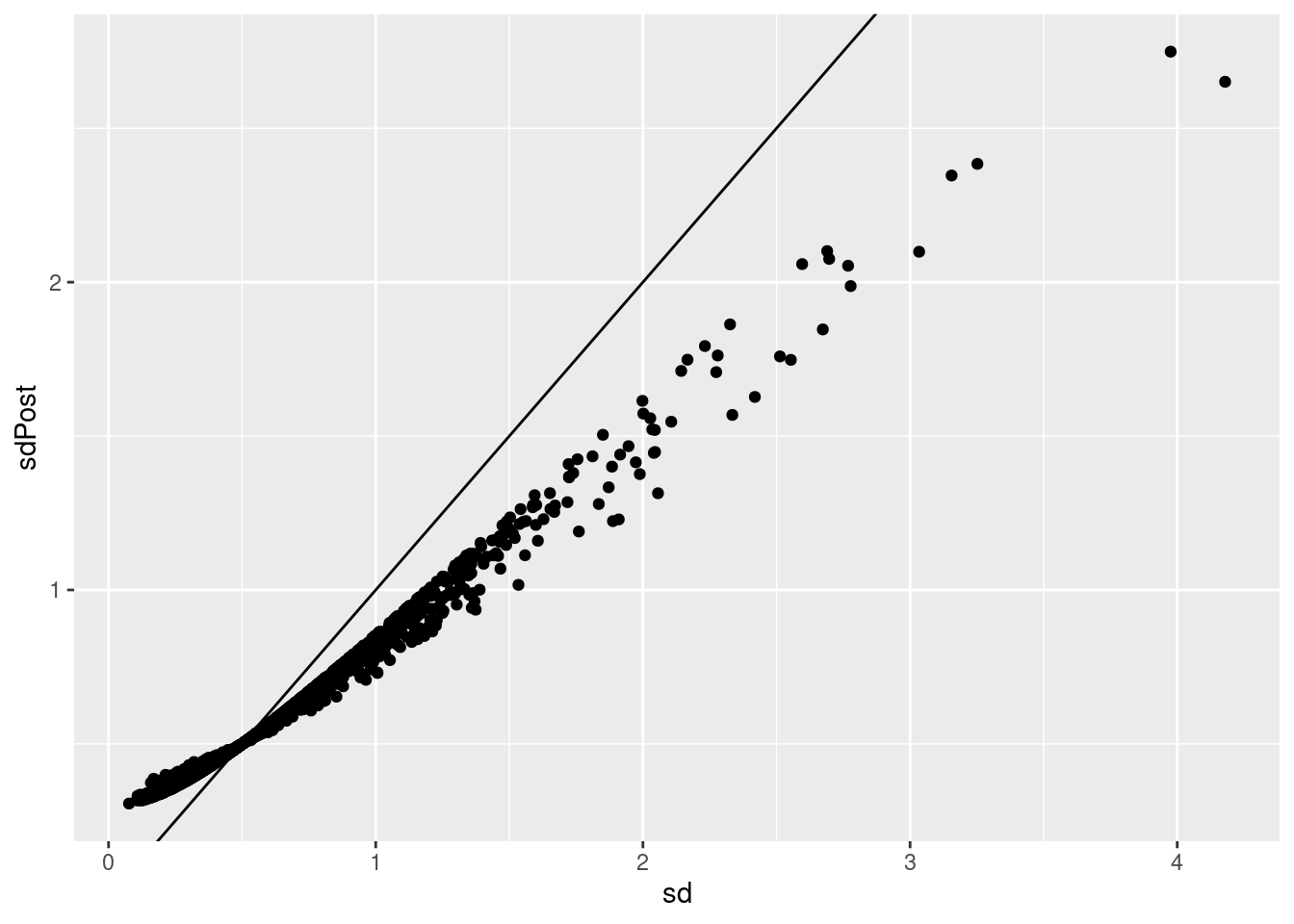
4.5.1 How do we get to the posterior standard deviation?
hlp <- limma::squeezeVar(
var = sapply(rowData(pe[["proteinRobust"]])$msqrobModels, getVar),
df = sapply(rowData(pe[["proteinRobust"]])$msqrobModels, getDF)
)4.5.1.1 Degrees of freedom of prior
hlp$df.prior## [1] 3.385413model <- rowData(pe[["proteinRobust"]])$msqrobModels[[2]]
getDfPosterior(model) - getDF(model)## [1] 3.3854134.5.1.2 posterior variance
\[\tilde s_p=\sqrt{\frac{d_p\times s^2_p + d_0 s_0^2}{d_p+d_0}} \]
hlp$var.prior## [1] 0.2489859varPost <- (getVar(model) * getDF(model) + hlp$df.prior * hlp$var.prior)/(getDF(model)+hlp$df.prior)
sqrt(varPost)## [1] 0.6607153getSigmaPosterior(model)## [1] 0.6607153Hence, standard deviations are shrunken towards prior standard deviation! Large standard deviations become smaller and smaller standard deviations become larger!
p1 +
geom_hline(yintercept = hlp$var.prior^.5)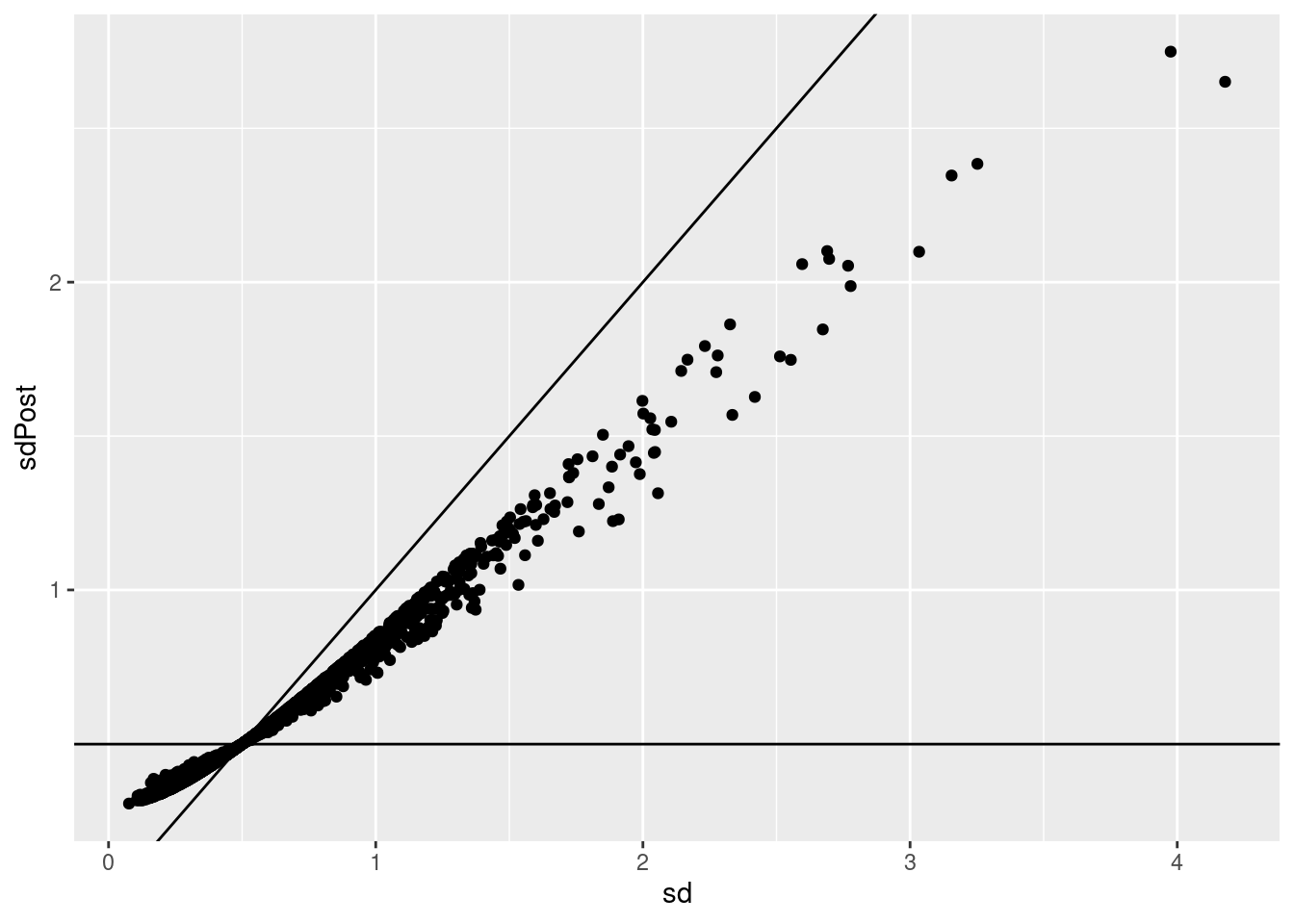
4.5.2 Illustration via Simulation
Suppose that the standard deviations for all proteins are the same and are equal to 1. We simulate proteins with the same mean as the fitted mean in the experiment but with standard deviation of 1.
nCoefs <- getCoef(rowData(pe[["proteinRobust"]])$msqrobModels[[2]]) %>% length
coefs <-
sapply(rowData(pe[["proteinRobust"]])$msqrobModels,
function(x) getCoef(x)[1:nCoefs]
) %>%
t %>%
na.exclude
p <- nrow(coefs)
n <- ncol(pe[[1]])
f0_equalVar <- sapply(1:p,
FUN=function(i, n, betas, sd, design) {
rnorm(n, mean = design %*% betas[i,], sd = sd)},
n = n,
betas = coefs,
sd = 1,
design = X
) %>%
t
colnames(f0_equalVar) <- colnames(pe[[1]])
sims <- readQFeatures(f0_equalVar %>% as.data.frame, ecol = 1:n, name = "sim_equalVar")
colData(sims) <- colData(pe)
sims <- msqrob(object = sims, i = "sim_equalVar", formula = ~ location*tissue + patient)
sd0 <- sapply(
rowData(sims[["sim_equalVar"]])$msqrobModels,
getSigma) %>%
na.exclude
sdPost0 <- sapply(
rowData(sims[["sim_equalVar"]])$msqrobModels,
getSigmaPosterior) %>%
na.exclude
qplot(sd0,sdPost0) +
geom_abline() +
ylim(range(sd0))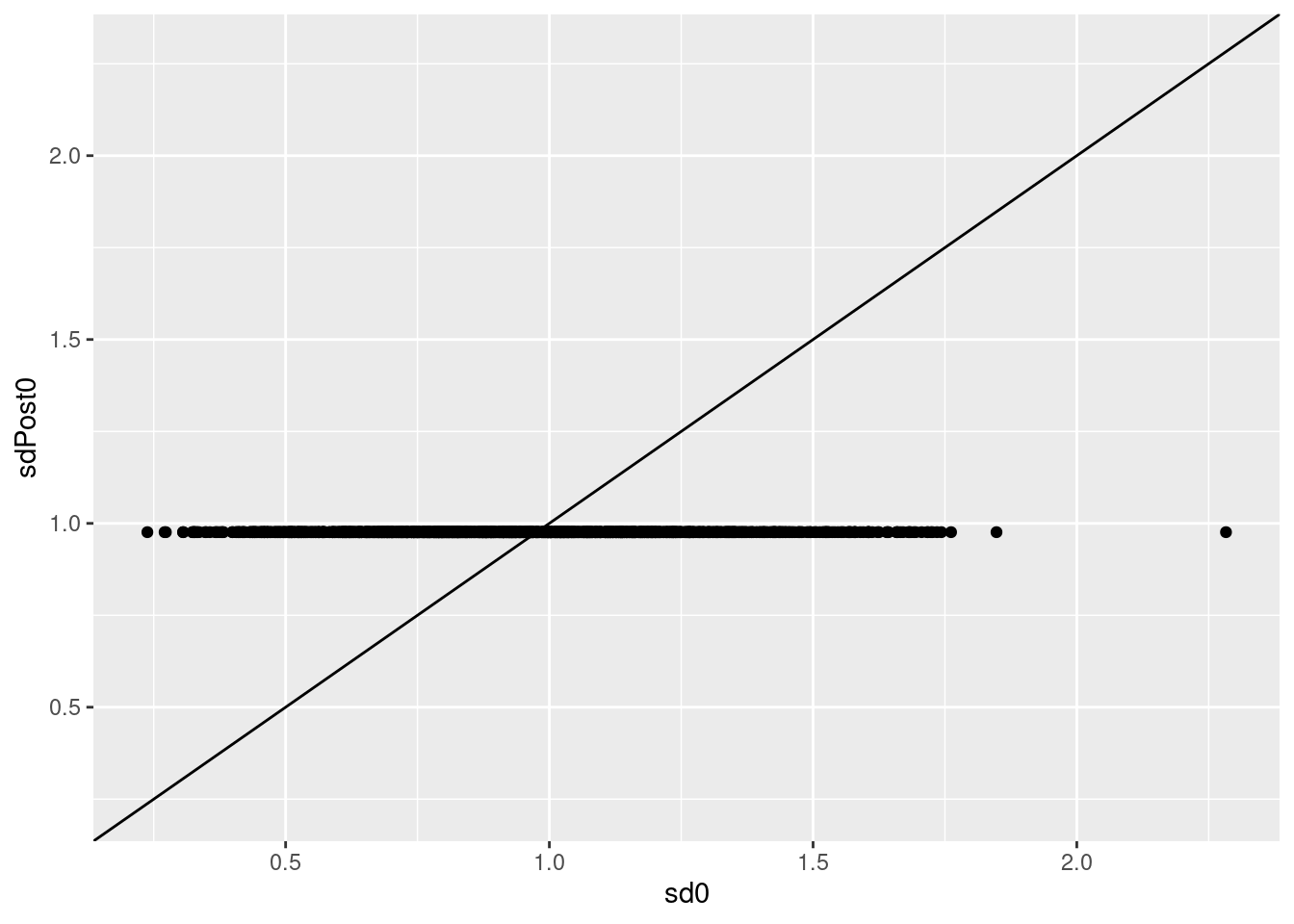
- We observe a large variability in the individual protein level standard deviation estimates.
- We simulated proteins with standard deviation of 1, but the protein estimates vary from 0.24, … , 2.28.
- Large uncertainty on the estimation of variances in small samples
- The empirical Bayes method, however, recognises that all proteins are simulated with the same variance.
- Hence, it can borrow tremendous strength across proteins to stabilize the variance estimation
- Here, it shrinks all protein variance to the prior variance, which is indeed very close to 1, the value we have adopted in the simulation.
Note, that the prior degrees of freedom are also set to infinity:
getDF(rowData(sims[["sim_equalVar"]])$msqrobModels[[1]])## [1] 5.911521getDfPosterior(rowData(sims[["sim_equalVar"]])$msqrobModels[[1]])## [1] Infwhich imposes shrinkage to the prior standard deviation!
The empirical Bayes method can thus indeed recognize the common variance that is shared across all simulated proteins!
5 P-values
5.1 Simulation under H_0.
- Mean log2 protein intensity for atrium equals mean log2 protein intensity for ventriculum in the left heart region.
- sd equals the sd for the protein.
- Extract \(\hat \sigma\) and \(\beta\)’s
sd <- sapply(
rowData(pe[["proteinRobust"]])$msqrobModels,
getSigma) %>%
na.exclude
coefs <-
sapply(rowData(pe[["proteinRobust"]])$msqrobModels,
function(x) getCoef(x)[1:nCoefs]
) %>%
t %>%
na.exclude- Set \(\beta_\text{tissue}\) equal to 0. No FC between atrium and ventriculum left.
coefs0 <- coefs
coefs0[,3] <- 0- Simulate protein expressions for each protein from a Normal distribution under \(H_0\) for left heart region (no FC between atrium and ventriculum left) and sd the sd for the protein.
set.seed(104)
f0 <- sapply(1:p,
function(i, betas, sd, design)
rnorm(n, mean = design %*% betas[i,], sd = sd[i]),
betas = coefs0,
sd = sd,
design = X
) %>%
t
colnames(f0) <- colnames(pe[[1]])- Setup QFeatures object and perform MSqRob analysis
sims <- readQFeatures(f0 %>% as.data.frame, ecol = 1:n, name = "sim0")
colData(sims) <- colData(pe)
sims <- msqrob(object = sims, i = "sim0", formula = ~ location*tissue + patient)
sims <- hypothesisTest(object = sims, i = "sim0", contrast = L)5.1.1 Evaluate pvalues under H_0
volcano <- ggplot(rowData(sims[["sim0"]])$tissueV,
aes(x = logFC, y = -log10(pval), color = pval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) + theme_minimal()
volcano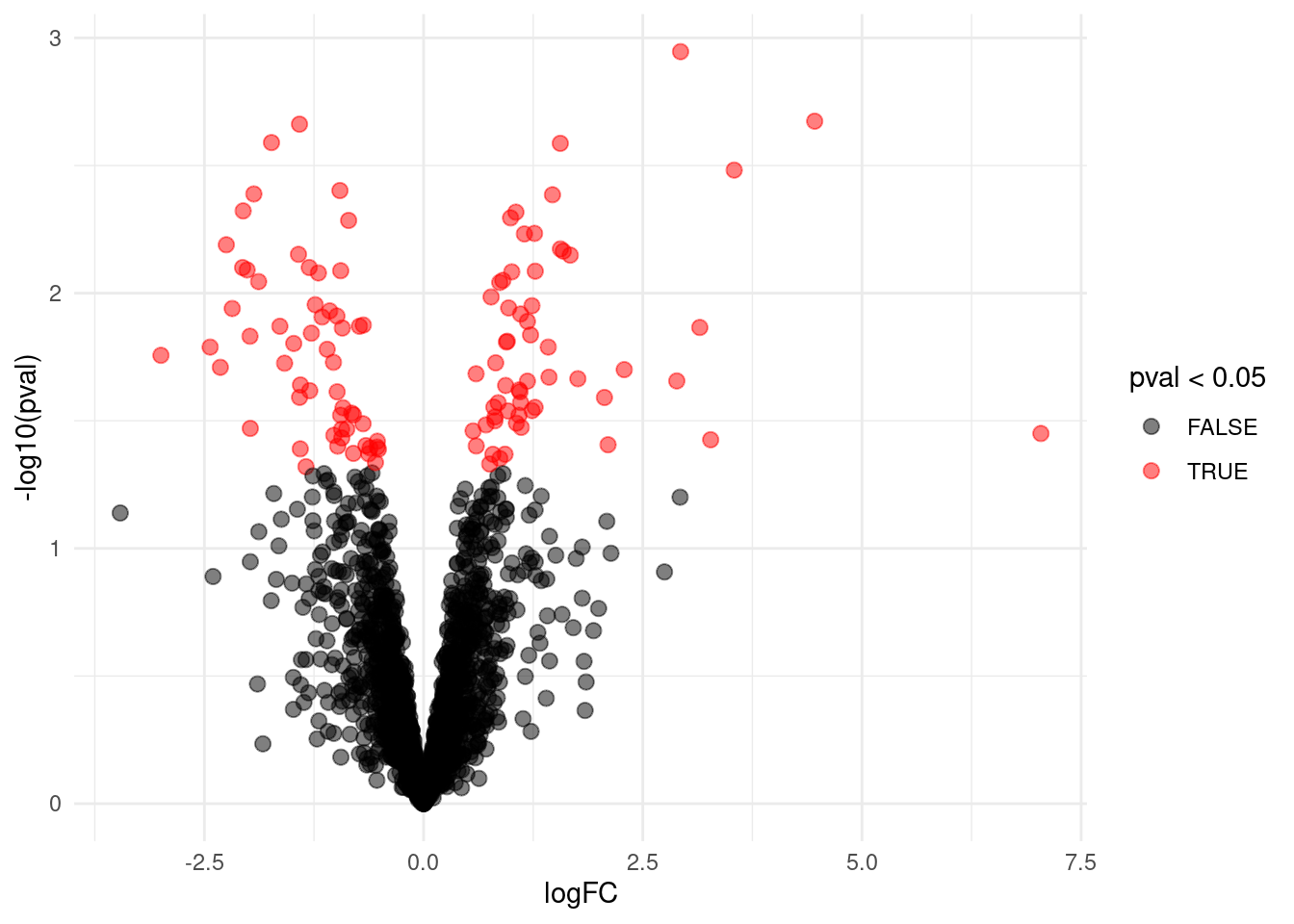
Number of false positives without multiple testing?
rowData(sims[["sim0"]])$tissueV %>%
filter(pval <0.05) %>%
nrow## [1] 115mean(rowData(sims[["sim0"]])$tissueV$pval < 0.05) ## [1] 0.05665025hist(rowData(sims[["sim0"]])$tissueV$pval,main = "simulation H0")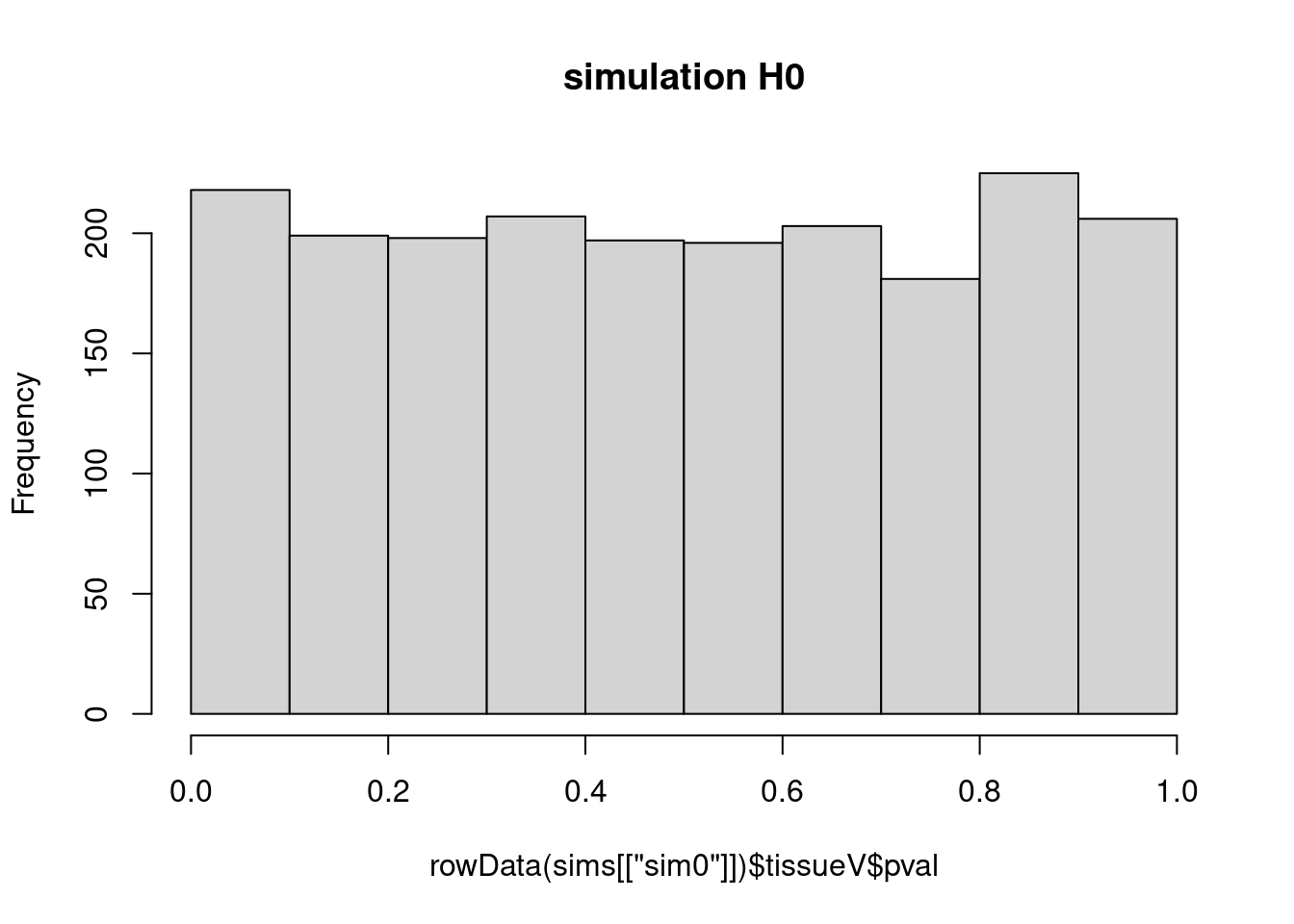
- The p-values are uniform!
- All p-values under the null are equally likely.
- Statistical hypthesis testing leads to a uniform test strategy under \(H_0\)
- If use p-value cutoff at 0.05 we expect to return 5% of the non-DE proteins as differentially expressed: many false positives can be expected!
5.2 Pvalue distribution in real experiment
hist(rowData(pe[["proteinRobust"]])$tissueV$pval, main = "realData")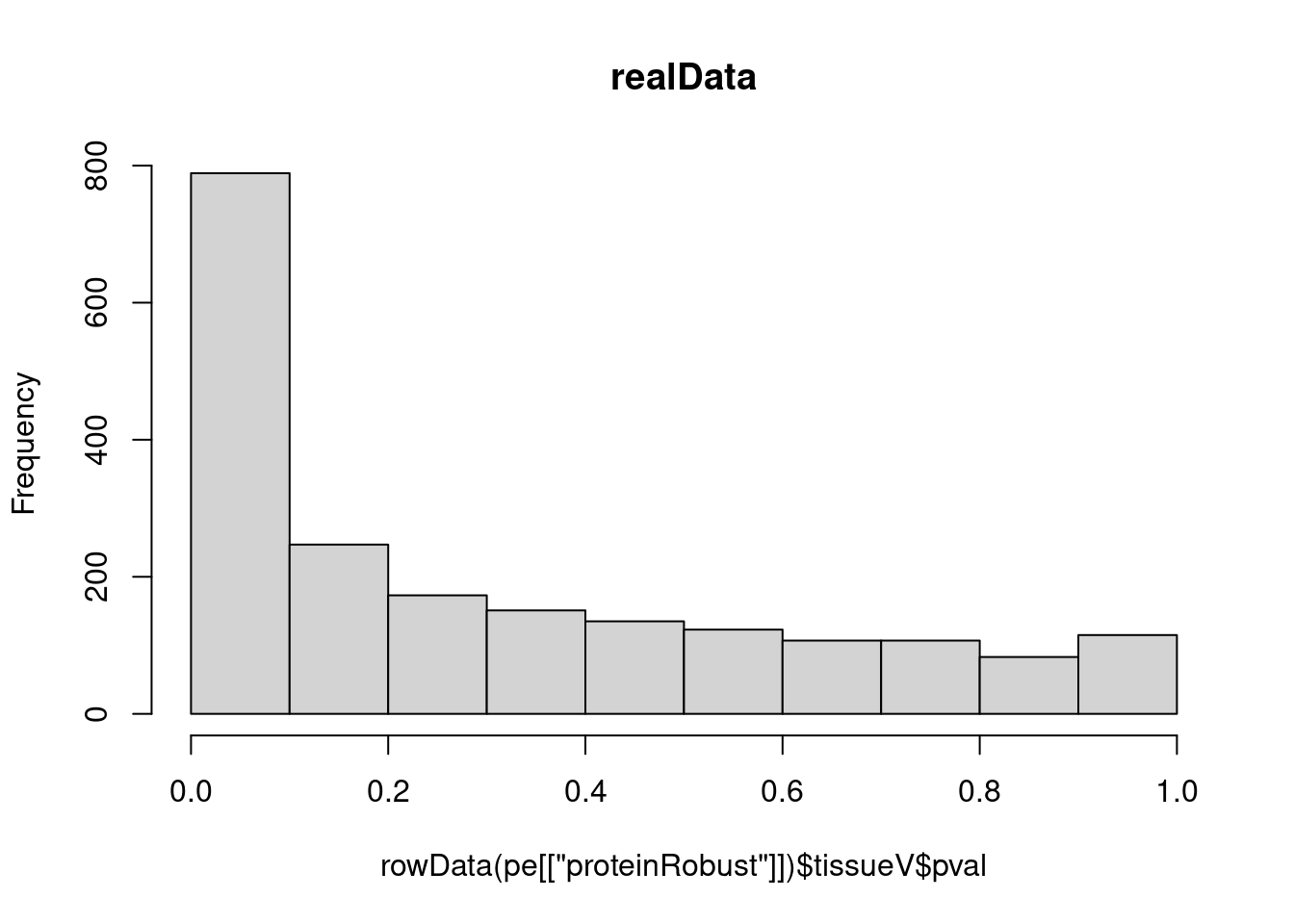
- A mixture of null proteins (non-DE): uniform, and, DE proteins: enrichment of p-values at low p-values
6 Correction for multiple testing
- We can adjust the p-values for multiple testing.
6.1 Family wise error rate correction:
A list of returned proteins is considered to be in error as soon as it contains at most one false positive protein.
\(\text{FWER} = P(FP \leq 1)\)
FWER: probability of making at least one false positive decision or probability to declare at least one protein differentially abundant which is truly non differentially abundant
6.1.1 Bonferroni method
- Simple method
- \(m\) tests are performed at the level \(\alpha/m\)
- FWER\(\leq\sum\limits_{p=1}^{m}P(reject H_{0p}\vert H_{0p}\text{ is true})=m \alpha/m=\alpha\)
- Provides strong control
- Bonferroni is very conservative
- Works for dependent tests
- Adjusted p-value: \(\tilde{p}_p=\min(m\ p_p,1)\)
6.1.2 Bonferroni in practise
Via R functions
padj <- p.adjust(
rowData(pe[["proteinRobust"]])$tissueV$pval,
method = "bonferroni")Own Implementation: adjust and make sure that p-value is smaller than 1.
m <- sum(!is.na(rowData(pe[["proteinRobust"]])$tissueV$pval))
padjSelf <- rowData(pe[["proteinRobust"]])$tissueV$pval * m
padjSelf[padjSelf > 1] <- 1
range(padj - padjSelf, na.rm = TRUE)## [1] 0 06.1.3 Illustration in simulation under \(H_0\) and heart case study
volcano <- ggplot(rowData(sims[["sim0"]])$tissueV,
aes(x = logFC, y = -log10(pval), color = p.adjust(pval,"bonferroni") < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) +
theme_minimal() +
ggtitle("simulated heart data under H0")
volcano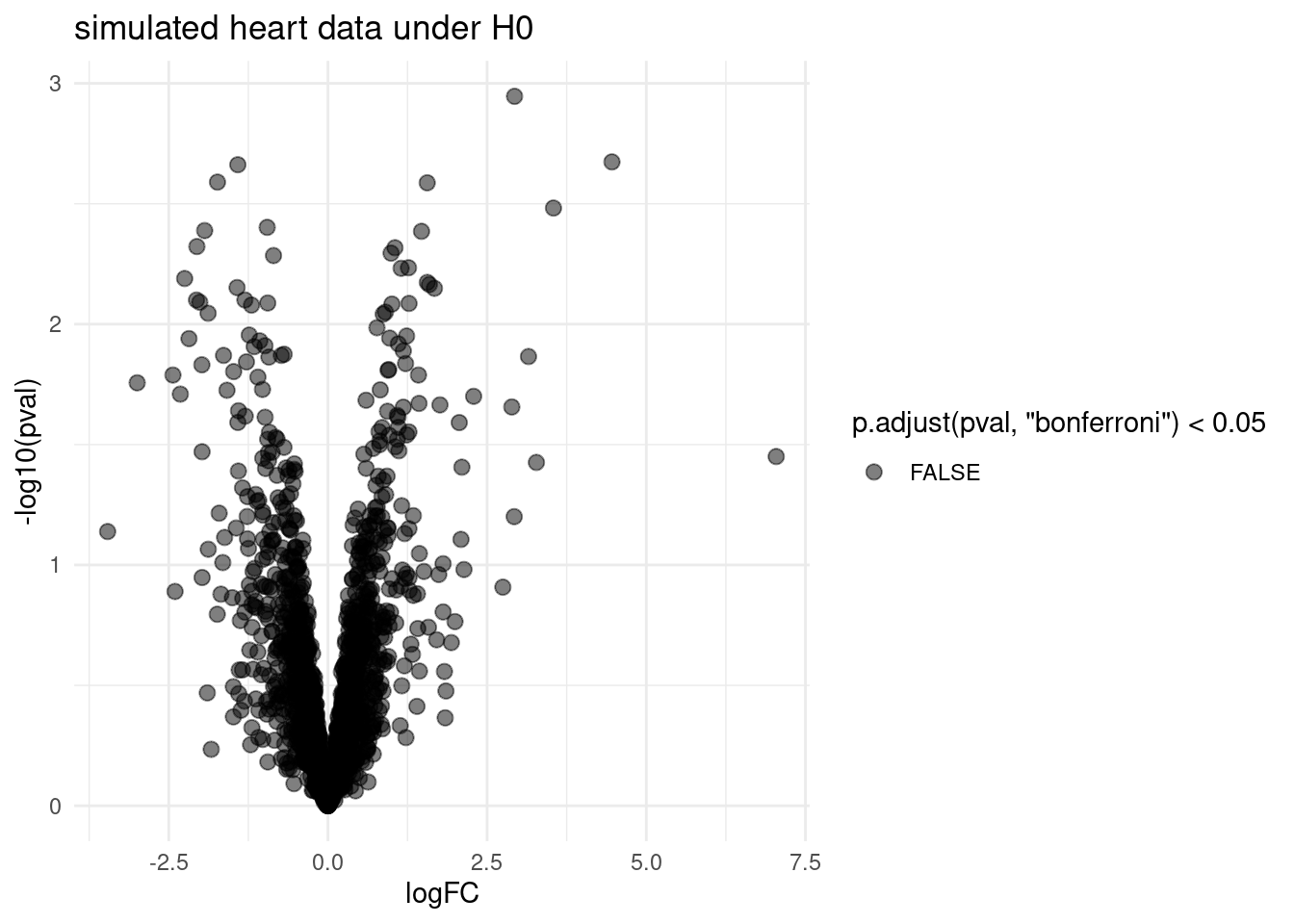
- No false positives are returned for simulation under H_0. List is correct according to FWER.
volcano <- ggplot(rowData(pe[["proteinRobust"]])$tissueV,
aes(x = logFC, y = -log10(pval), color = p.adjust(pval,"bonferroni") < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) +
theme_minimal() +
ggtitle("real heart data")
volcano## Warning: Removed 1263 rows containing missing values (geom_point).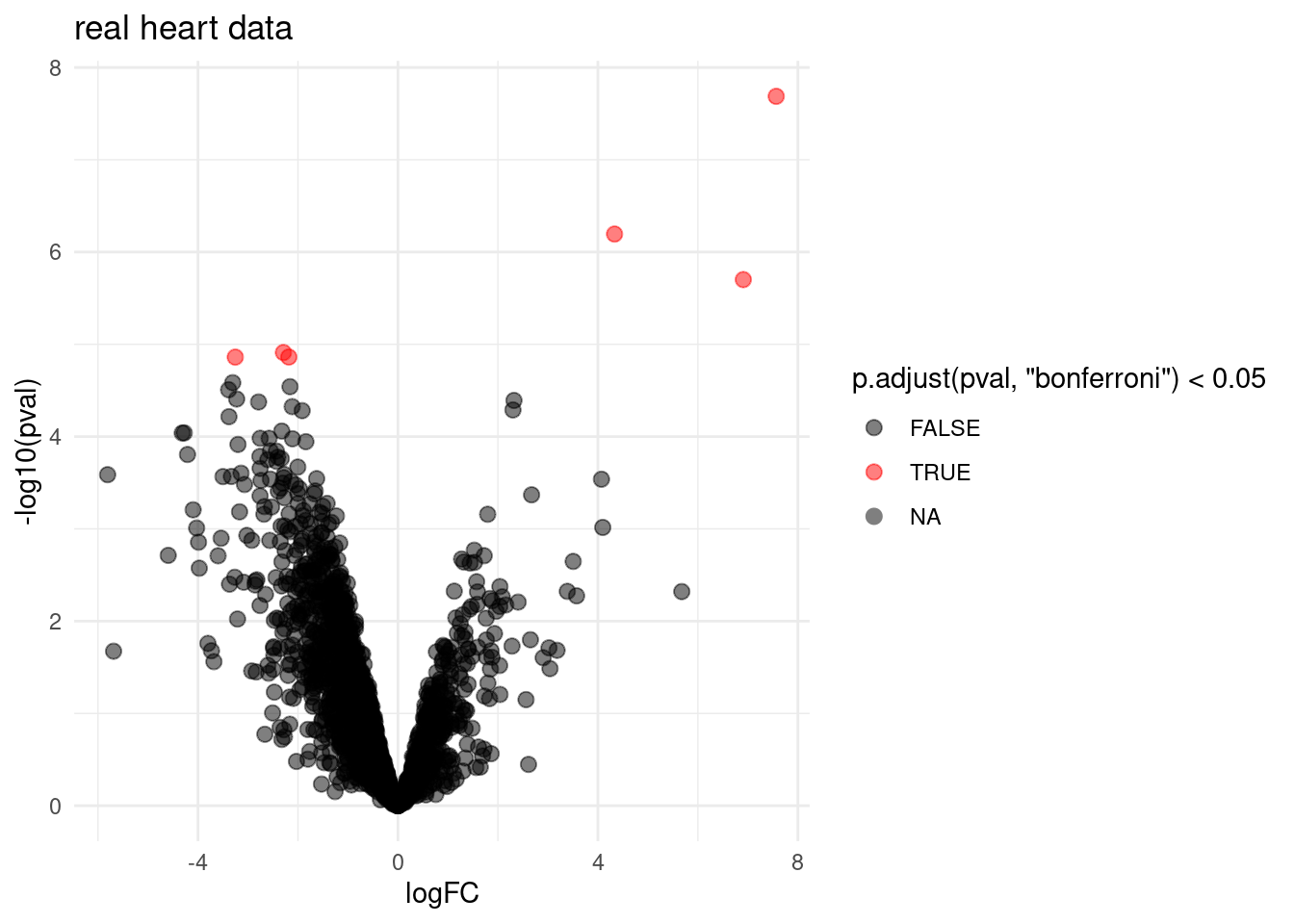
- Very few proteins are returned for real data. Very conservative!
6.2 False discovery rate
- Adjusted P-values with the Benjamini Hochberg correction correspond to the estimated FDR of the set that is returned when the significance level is set at this threshold. \[\begin{eqnarray} FDR(p_0) &=& \text{E}\left[\frac{FP}{(FP + TP)}\right]\\ &\approx&\frac{p_0 \times m}{\#p_p \leq p_0}\\ \end{eqnarray}\]
So adjusted p-value for protein j equals \[\tilde p_j = \frac{p_{0,j} \times m}{\#p_p \leq p_{0,j}}\]
However, the FDR always has to be between 0 and 1 so:
\[\tilde p_j = \min\left[\frac{p_{0,j} \times m}{\#p_p \leq p_{0,j}},1\right]\]
and the adjusted p-values should remain in the same order as the original p-values.
\[\tilde p_j = \min\limits_{\forall k: p_k > p_j} \min\left[\frac{p_{0,k} \times m}{\#p_p \leq p_{0,k}},1\right]\]
- Order pvalues
pvals <- rowData(pe[["proteinRobust"]])$tissueV$pval
naInd <- is.na(pvals)
pHlp <- pvals[!naInd]
ord <- pHlp %>% order
pHlp <- pHlp[ord]- Adjust ordered p-values
pHlp <- pHlp*length(pHlp)/(1:length(pHlp))- Ensure adjust p-values are smaller are equal than 1
pHlp[pHlp>1] <- 1- Monotonicity constraint
pmin <- pHlp[length(pHlp)]
for (j in (length(pHlp)-1):1)
{
if (pHlp[j] < pmin)
pmin <- pHlp[j] else
pHlp[j] <- pmin
}- Put p-values back in original order
pHlp[ord] <- pHlp
pAdj <- pvals
pAdj[!naInd] <- pHlp
head(pAdj)## [1] NA 0.8351039 NA NA 0.9062353 NAhead(rowData(pe[["proteinRobust"]])$tissueV)range(rowData(pe[["proteinRobust"]])$tissueV$adjPval - pAdj,na.rm=TRUE)## [1] -2.220446e-16 2.220446e-166.2.1 Illustration in simulation under \(H_0\) and heart case study
volcano <- ggplot(rowData(sims[["sim0"]])$tissueV,
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) +
theme_minimal() +
ggtitle("simulated heart data under H0")
volcano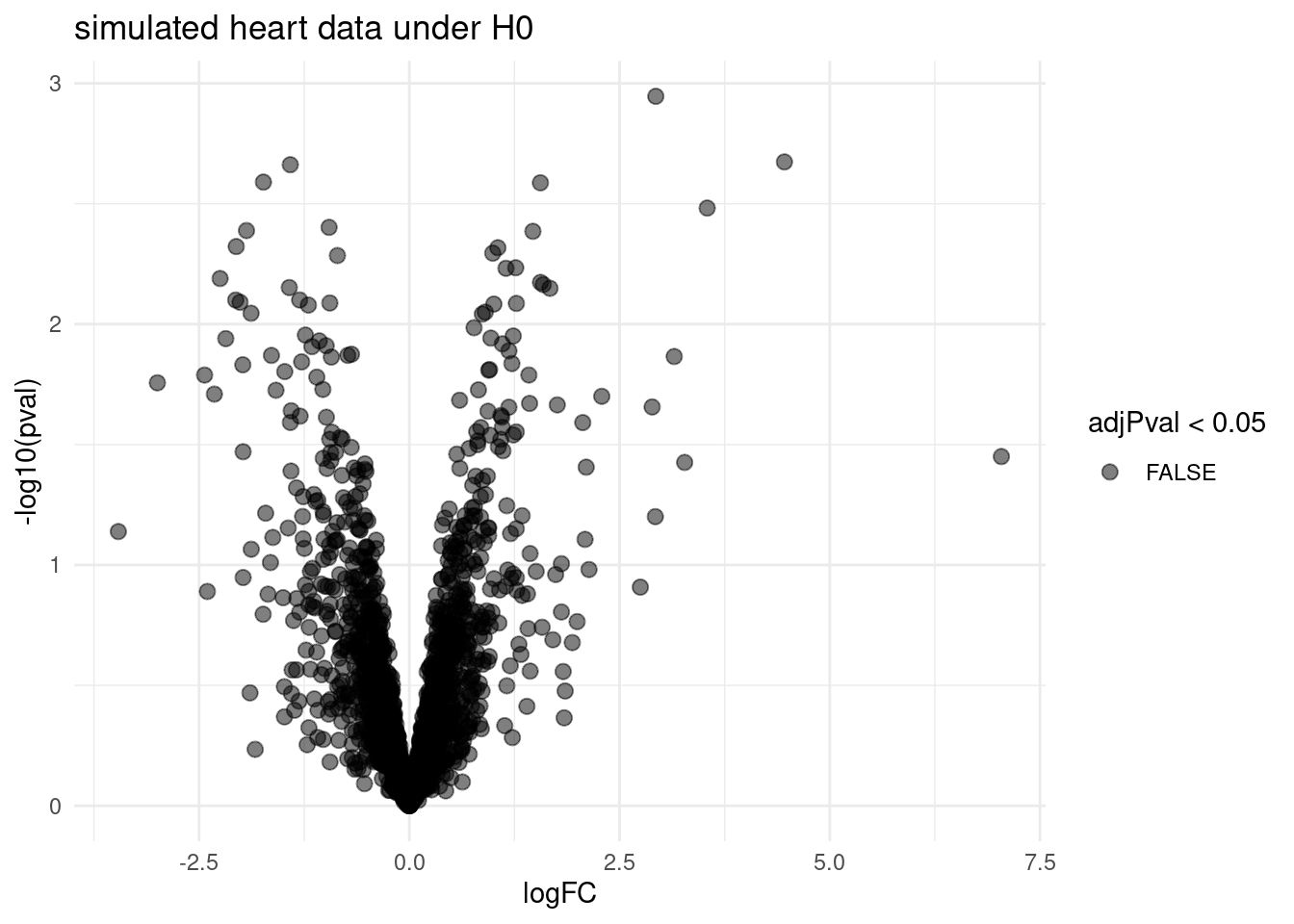
- No false positives are returned for simulation under H_0. List is correct according to FWER.
- It can be shown that the FDR-method controls the FWER when \(H_0\) is true for all features.
volcano <- ggplot(rowData(pe[["proteinRobust"]])$tissueV,
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) +
theme_minimal() +
ggtitle("real heart data")
volcano## Warning: Removed 1263 rows containing missing values (geom_point).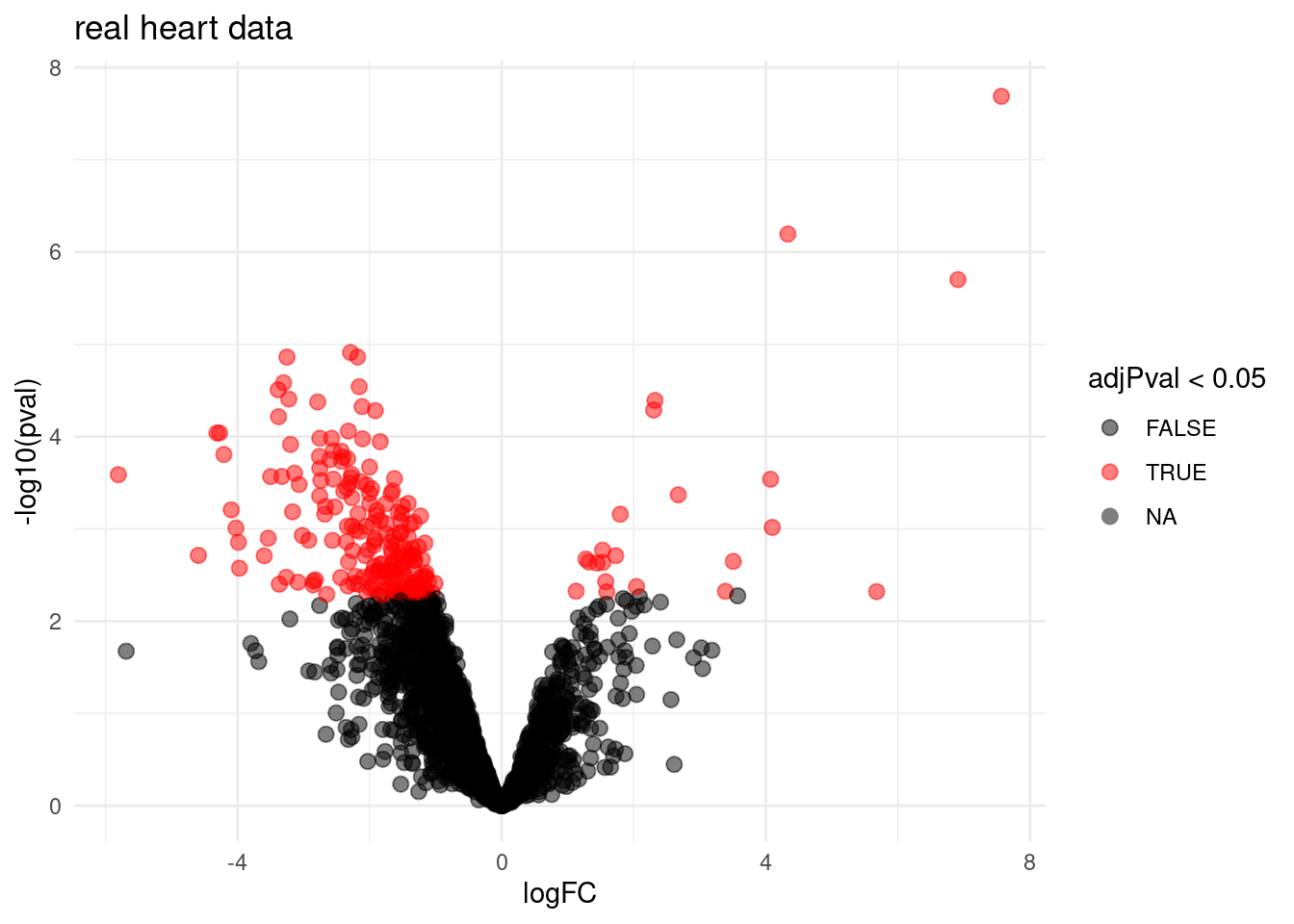
The FDR method allows us to return much longer DA protein lists at the expense of a few false positives. The FDR controls the fraction of false positives in the list that you return on average on the significance level that is adopted. So if you use \(\alpha=0.05\) we expect on average 5% of false positives in the list that we return.