Proteomics data analysis: heart
Lieven Clement
statOmics, Ghent University (https://statomics.github.io)
1 Background
Researchers have assessed the proteome in different regions of the heart for 3 patients (identifiers 3, 4, and 8). For each patient they sampled the left atrium (LA), right atrium (RA), left ventricle (LV) and the right ventricle (RV). The data are a small subset of the public dataset PXD006675 on PRIDE.
Suppose that researchers are mainly interested in comparing the ventricular to the atrial proteome. Particularly, they would like to compare the left atrium to the left ventricle, the right atrium to the right ventricle, the average ventricular vs atrial proteome and if ventricular vs atrial proteome shifts differ between left and right heart region.
2 Data
We first import the peptides.txt file. This is the file that contains your peptide-level intensities. For a MaxQuant search [6], this peptides.txt file can be found by default in the “path_to_raw_files/combined/txt/” folder from the MaxQuant output, with “path_to_raw_files” the folder where raw files were saved. In this tutorial, we will use a MaxQuant peptides file from MaxQuant that can be found in the data tree of the SGA2020 github repository https://github.com/statOmics/SGA2020/tree/data/quantification/heart .
To import the data we use the QFeatures package.
We generate the object peptideRawFile with the path to the peptideRaws.txt file. Using the grepEcols function, we find the columns that contain the expression data of the peptideRaws in the peptideRaws.txt file.
library(tidyverse)
library(limma)
library(QFeatures)
library(msqrob2)
library(plotly)
peptidesFile <- "https://raw.githubusercontent.com/statOmics/SGA2020/data/quantification/heart/peptides.txt"
ecols <- MSnbase::grepEcols(
peptidesFile,
"Intensity ",
split = "\t")
pe <- readQFeatures(
table = peptidesFile,
fnames = 1,
ecol = ecols,
name = "peptideRaw", sep="\t")
pe## An instance of class QFeatures containing 1 assays:
## [1] peptideRaw: SummarizedExperiment with 31319 rows and 12 columns## class: SummarizedExperiment
## dim: 31319 12
## metadata(0):
## assays(1): ''
## rownames(31319): AAAAAAAAAK AAAAAAAAEQQSSNGPVK ... YYTPVPCESATAK
## YYTYLIMNK
## rowData names(91): Sequence N.term.cleavage.window ...
## Oxidation..M..site.IDs MS.MS.Count
## colnames(12): Intensity.LA3 Intensity.LA4 ... Intensity.RV4
## Intensity.RV8
## colData names(0):We will make use from data wrangling functionalities from the tidyverse package. The %>% operator allows us to pipe the output of one function to the next function.
colData(pe)$location <- substr(
colnames(pe[["peptideRaw"]]),
11,
11) %>%
unlist %>%
as.factor
colData(pe)$tissue <- substr(
colnames(pe[["peptideRaw"]]),
12,
12) %>%
unlist %>%
as.factor
colData(pe)$patient <- substr(
colnames(pe[["peptideRaw"]]),
13,
13) %>%
unlist %>%
as.factorWe calculate how many non zero intensities we have per peptide and this will be useful for filtering.
Peptides with zero intensities are missing peptides and should be represent with a NA value rather than 0.
2.1 Data exploration
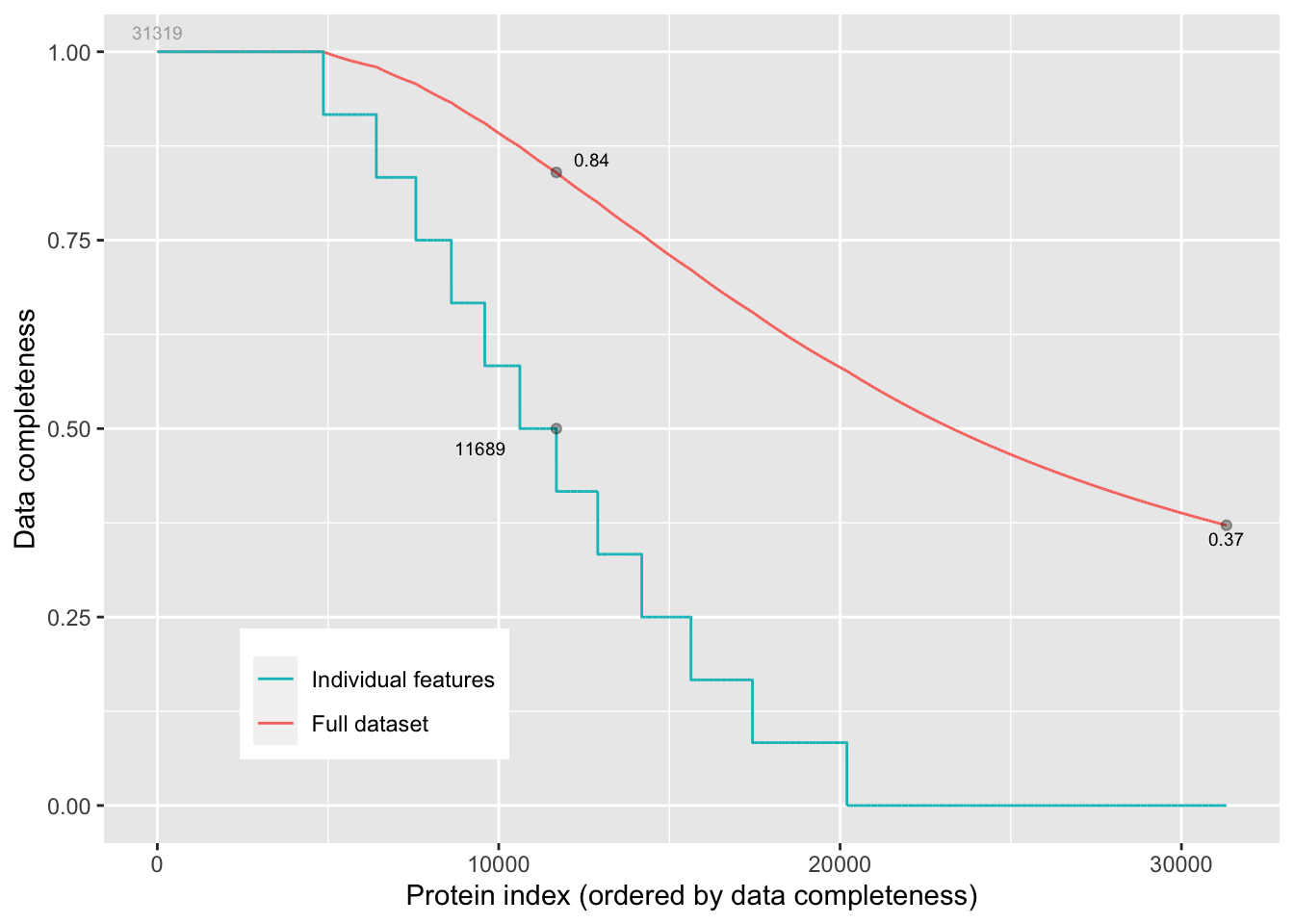
We can inspect the missingness in our data with the plotNA() function provided with MSnbase. 63% of all peptide intensities are missing and for some peptides we do not even measure a signal in any sample. The missingness is similar across samples.

3 Preprocessing
This section preforms standard preprocessing for the peptide data. This include log transformation, filtering and summarisation of the data.
3.1 Log transform the data
pe <- logTransform(pe, base = 2, i = "peptideRaw", name = "peptideLog")
limma::plotDensities(assay(pe[["peptideLog"]]))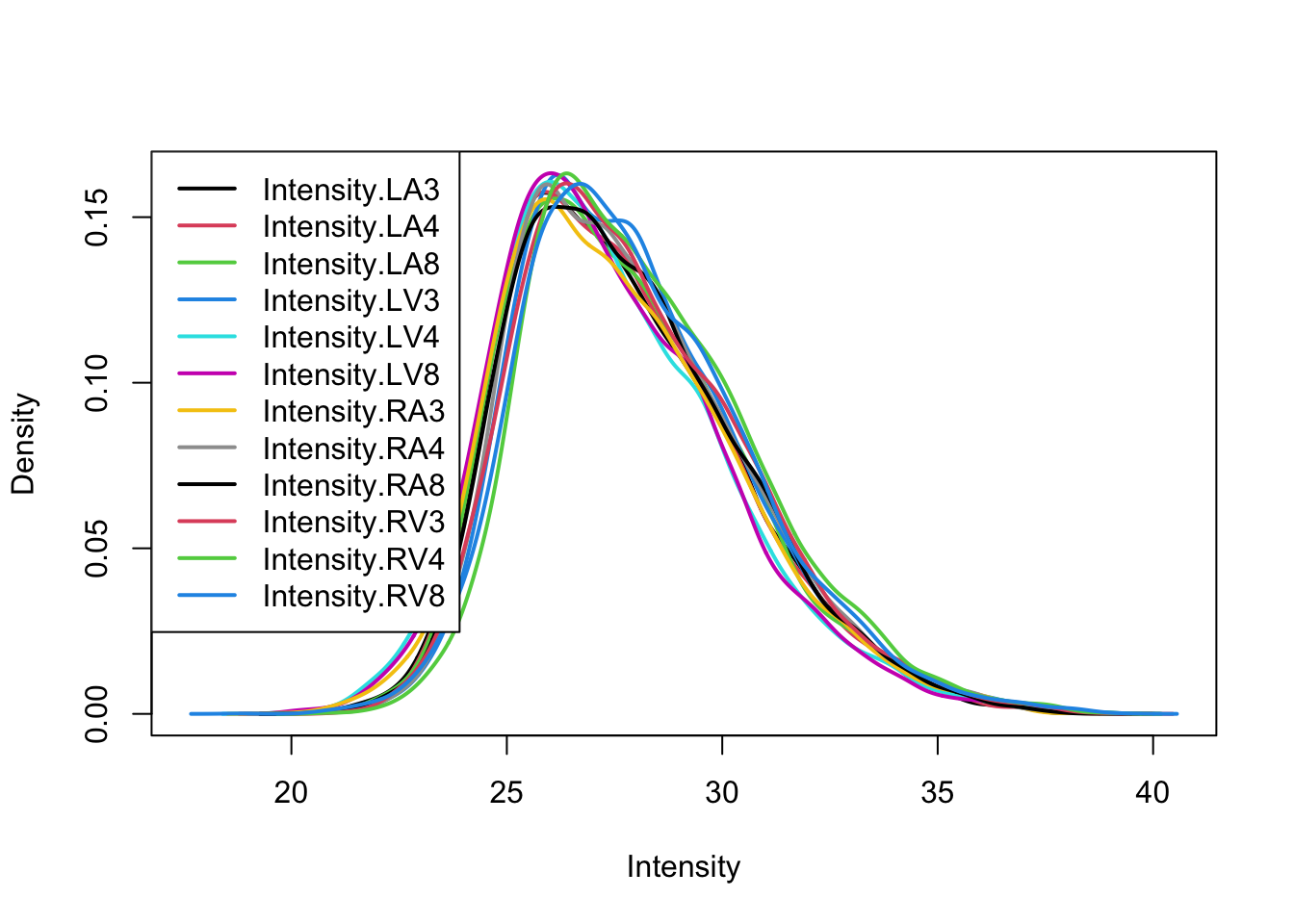
3.2 Filtering
3.2.1 Handling overlapping protein groups
In our approach a peptide can map to multiple proteins, as long as there is none of these proteins present in a smaller subgroup.
3.2.2 Remove reverse sequences (decoys) and contaminants
We now remove the contaminants, peptides that map to decoy sequences, and proteins which were only identified by peptides with modifications.
First look to the names of the variables for the peptide features
## [1] "Sequence" "N.term.cleavage.window"
## [3] "C.term.cleavage.window" "Amino.acid.before"
## [5] "First.amino.acid" "Second.amino.acid"
## [7] "Second.last.amino.acid" "Last.amino.acid"
## [9] "Amino.acid.after" "A.Count"
## [11] "R.Count" "N.Count"
## [13] "D.Count" "C.Count"
## [15] "Q.Count" "E.Count"
## [17] "G.Count" "H.Count"
## [19] "I.Count" "L.Count"
## [21] "K.Count" "M.Count"
## [23] "F.Count" "P.Count"
## [25] "S.Count" "T.Count"
## [27] "W.Count" "Y.Count"
## [29] "V.Count" "U.Count"
## [31] "O.Count" "Length"
## [33] "Missed.cleavages" "Mass"
## [35] "Proteins" "Leading.razor.protein"
## [37] "Start.position" "End.position"
## [39] "Gene.names" "Protein.names"
## [41] "Unique..Groups." "Unique..Proteins."
## [43] "Charges" "PEP"
## [45] "Score" "Identification.type.LA3"
## [47] "Identification.type.LA4" "Identification.type.LA8"
## [49] "Identification.type.LV3" "Identification.type.LV4"
## [51] "Identification.type.LV8" "Identification.type.RA3"
## [53] "Identification.type.RA4" "Identification.type.RA8"
## [55] "Identification.type.RV3" "Identification.type.RV4"
## [57] "Identification.type.RV8" "Fraction.Average"
## [59] "Fraction.Std..Dev." "Fraction.1"
## [61] "Fraction.2" "Fraction.3"
## [63] "Fraction.4" "Fraction.5"
## [65] "Fraction.6" "Fraction.7"
## [67] "Fraction.8" "Fraction.100"
## [69] "Experiment.LA3" "Experiment.LA4"
## [71] "Experiment.LA8" "Experiment.LV3"
## [73] "Experiment.LV4" "Experiment.LV8"
## [75] "Experiment.RA3" "Experiment.RA4"
## [77] "Experiment.RA8" "Experiment.RV3"
## [79] "Experiment.RV4" "Experiment.RV8"
## [81] "Intensity" "Reverse"
## [83] "Potential.contaminant" "id"
## [85] "Protein.group.IDs" "Mod..peptide.IDs"
## [87] "Evidence.IDs" "MS.MS.IDs"
## [89] "Best.MS.MS" "Oxidation..M..site.IDs"
## [91] "MS.MS.Count" "nNonZero"No information on decoys.
3.2.3 Remove peptides of proteins that were only identified with modified peptides
I will skip this step for the moment. Large protein groups file needed for this.
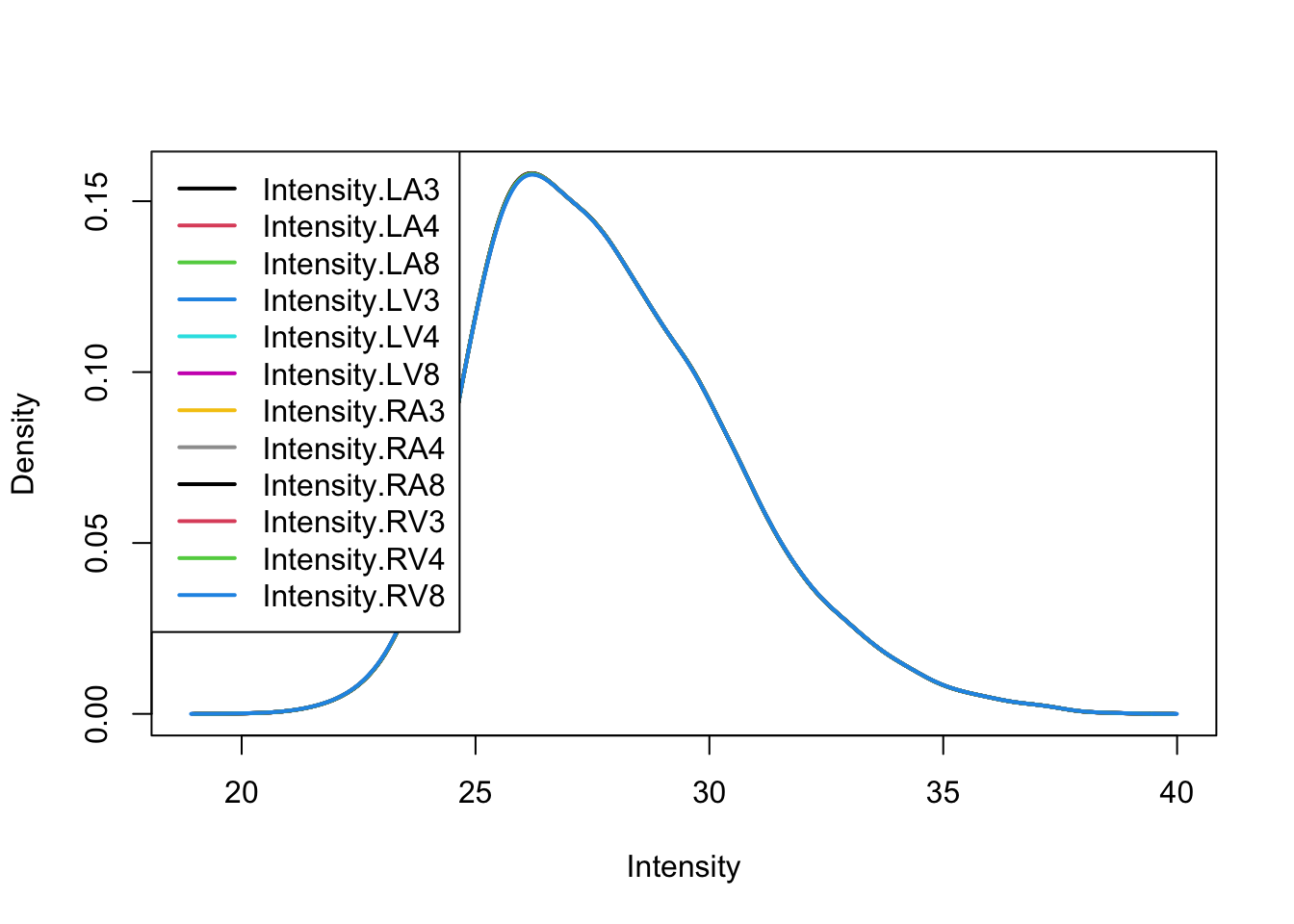
3.3 Quantile normalize the data
3.4 Explore quantile normalized data
After quantile normalisation the density curves for all samples coincide.
This is more clearly seen is a boxplot.
boxplot(assay(pe[["peptideNorm"]]), col = palette()[-1],
main = "Peptide distribtutions after normalisation", ylab = "intensity")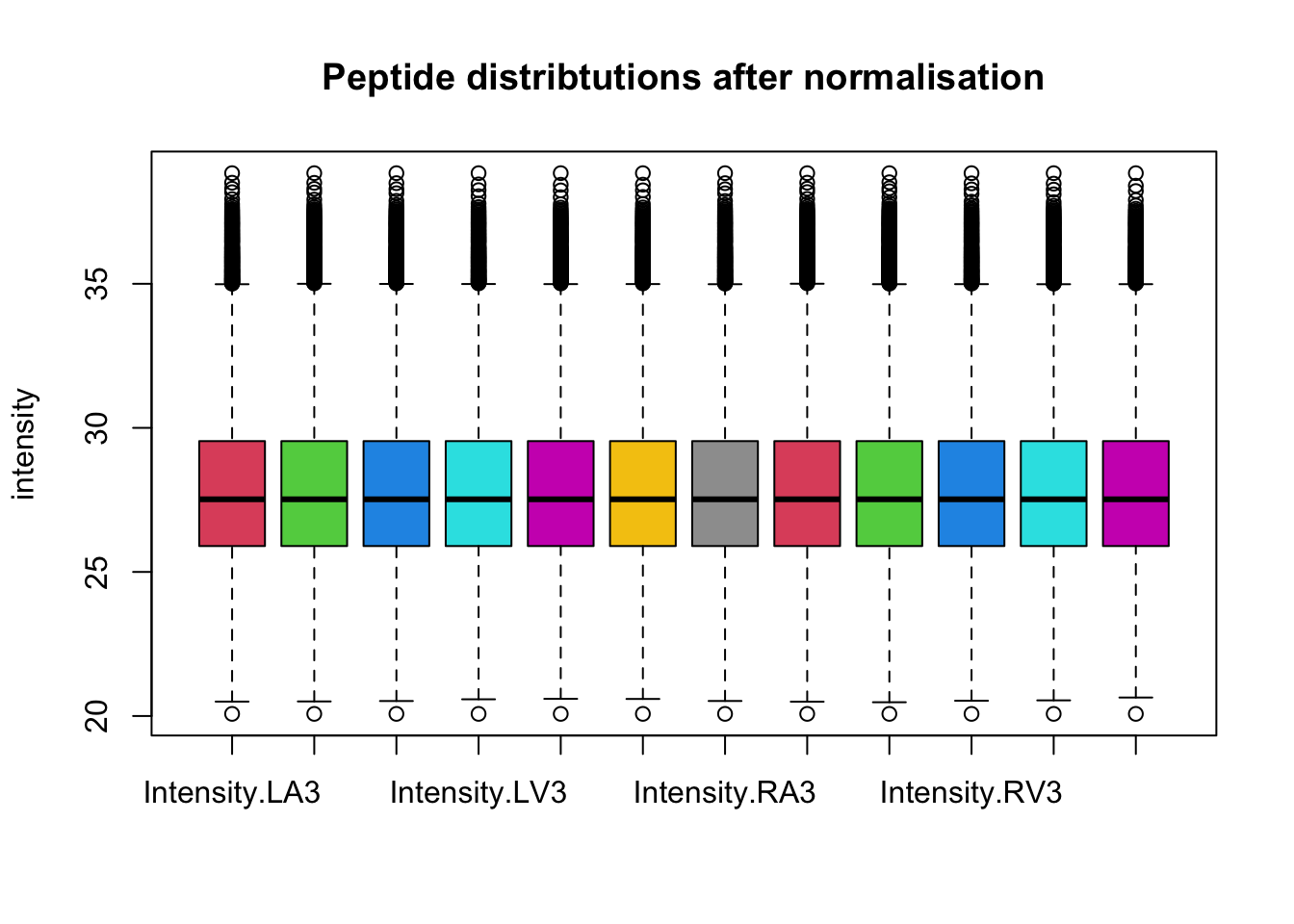
We can visualize our data using a Multi Dimensional Scaling plot, eg. as provided by the limma package.
limma::plotMDS(assay(pe[["peptideNorm"]]),
col = colData(pe)$location:colData(pe)$tissue %>%
as.numeric,
labels = colData(pe) %>%
rownames %>%
substr(start = 11, stop = 13)
)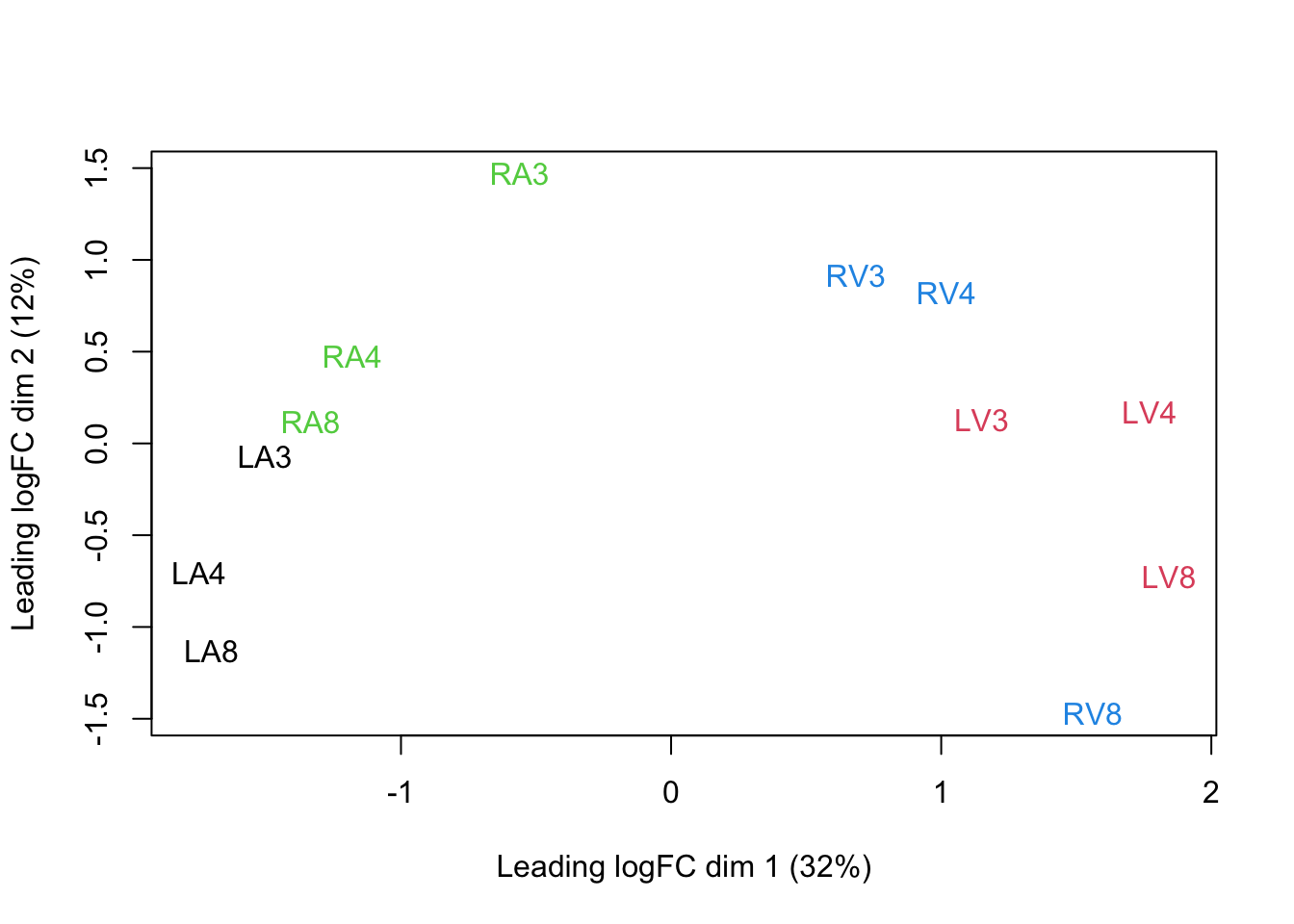
The first axis in the plot is showing the leading log fold changes (differences on the log scale) between the samples.
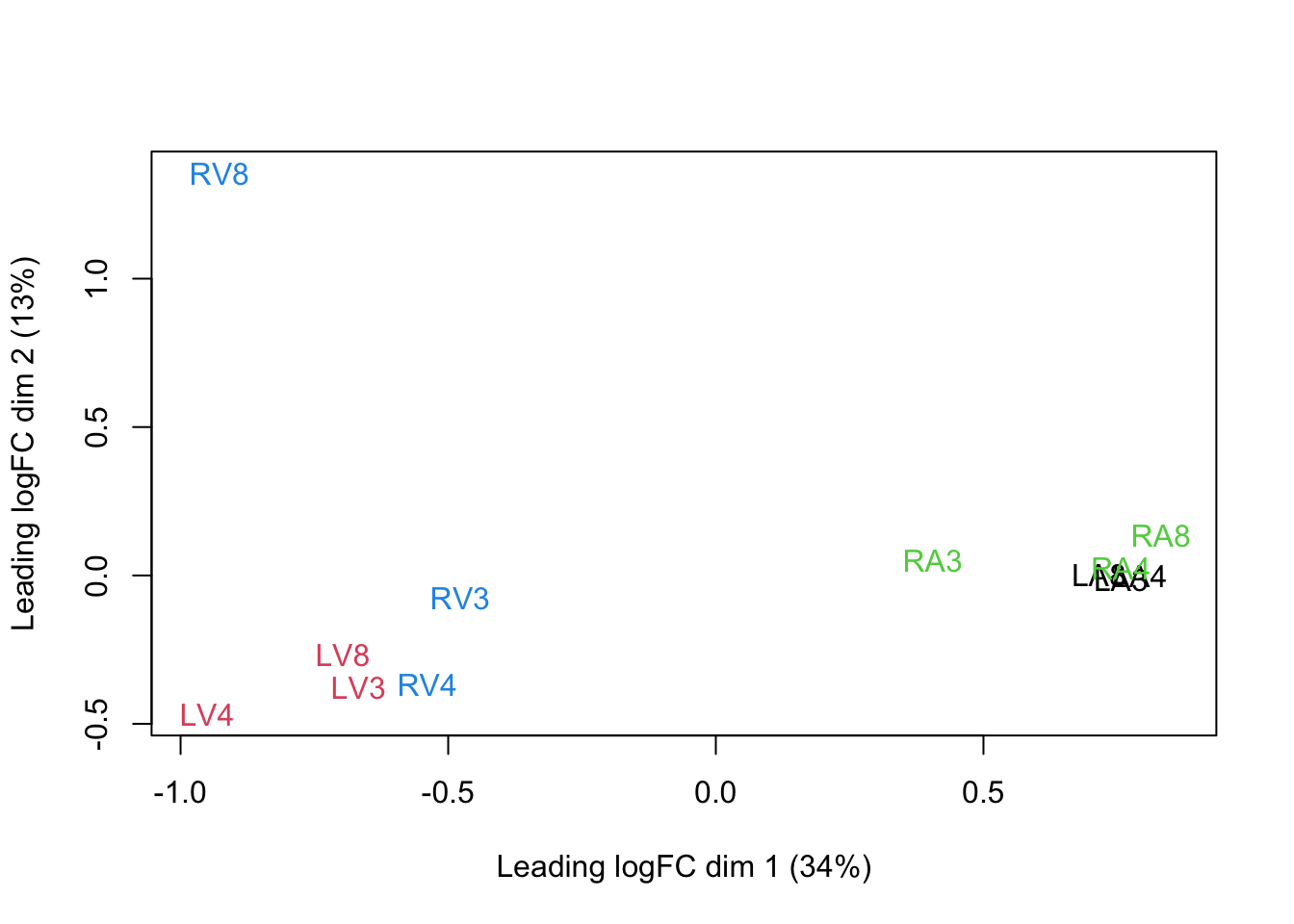
3.5 Summarization to protein level
We use robust summarization in aggregateFeatures. This is the default workflow of aggregateFeatures so you do not have to specifiy the argument fun. However, because we compare methods we have included the fun argument to show the summarization method explicitely.
pe <- aggregateFeatures(pe,
i = "peptideNorm",
fcol = "Proteins",
na.rm = TRUE,
name = "proteinRobust",
fun = MsCoreUtils::robustSummary)## Your quantitative and row data contain missing values. Please read the
## relevant section(s) in the aggregateFeatures manual page regarding the
## effects of missing values on data aggregation.plotMDS(assay(pe[["proteinRobust"]]),
col = colData(pe)$location:colData(pe)$tissue %>%
as.numeric,
labels = colData(pe) %>%
rownames %>%
substr(start = 11, stop = 13)
)
4 Data Analysis
4.1 Estimation
We model the protein level expression values using msqrob. By default msqrob2 estimates the model parameters using robust regression.
4.2 Inference
First, we extract the parameter names of the model.
## (Intercept) locationR tissueV patient4
## 26.33779939 0.28826213 0.20946392 0.46536955
## patient8 locationR:tissueV
## -0.02608046 -0.05550111L <- makeContrast(
c(
"tissueV = 0",
"tissueV + locationR:tissueV = 0",
"tissueV + 0.5*locationR:tissueV = 0","locationR:tissueV = 0"),
parameterNames =
rowData(pe[["proteinRobust"]])$msqrobModels[[2]] %>%
getCoef %>%
names
)
pe <- hypothesisTest(object = pe, i = "proteinRobust", contrast = L, overwrite=TRUE)4.3 Evaluate results contrast \(\log_2 FC_{V-A}^L\)
4.3.1 Volcano-plot
volcanoLeft <- ggplot(rowData(pe[["proteinRobust"]])$"tissueV + locationR:tissueV",
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) + theme_minimal()
volcanoLeft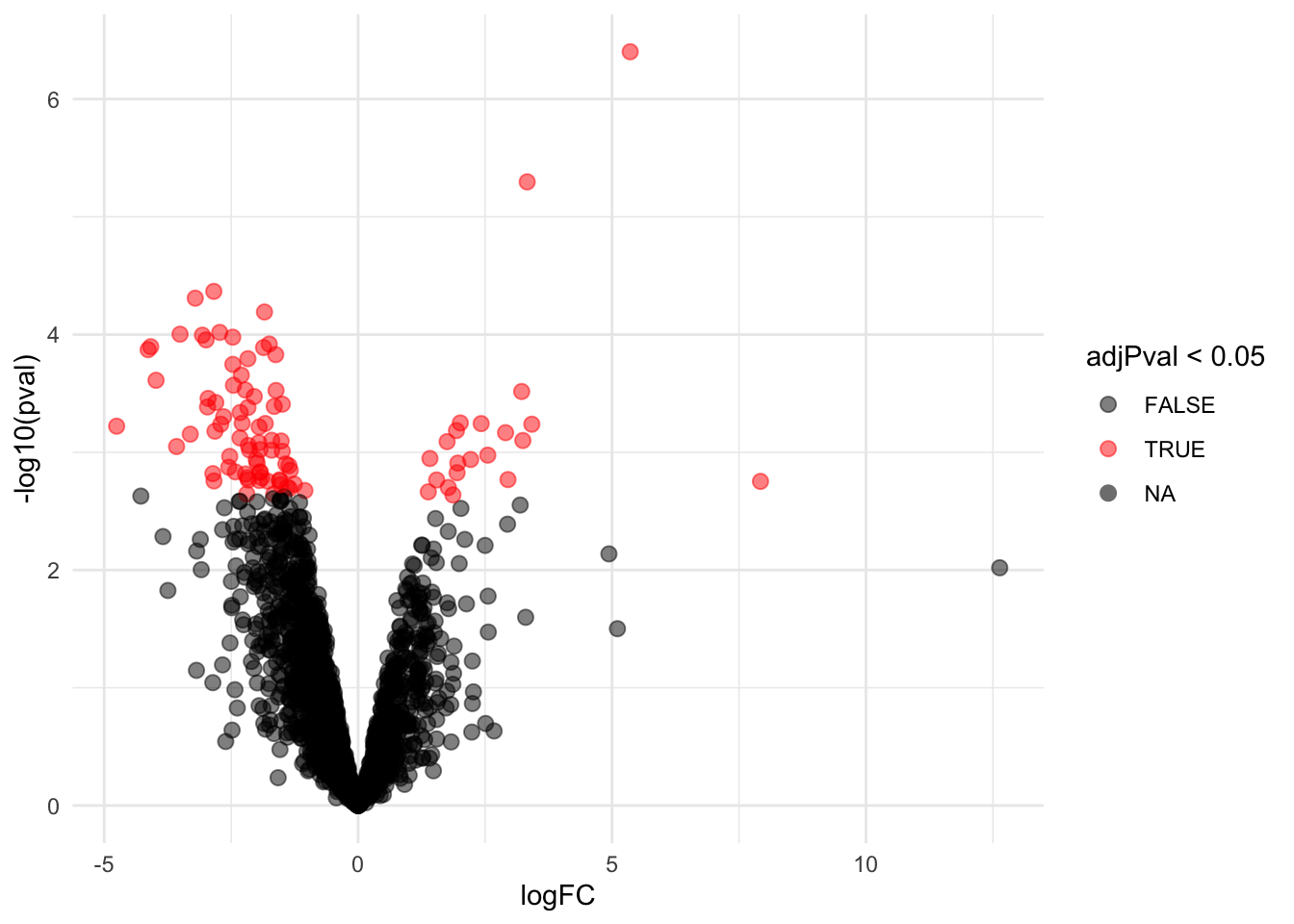
4.3.2 Heatmap
We first select the names of the proteins that were declared signficant.
sigNamesLeft <- rowData(pe[["proteinRobust"]])$tissueV %>%
rownames_to_column("proteinRobust") %>%
filter(adjPval<0.05) %>%
pull(proteinRobust)
heatmap(assay(pe[["proteinRobust"]])[sigNamesLeft, ])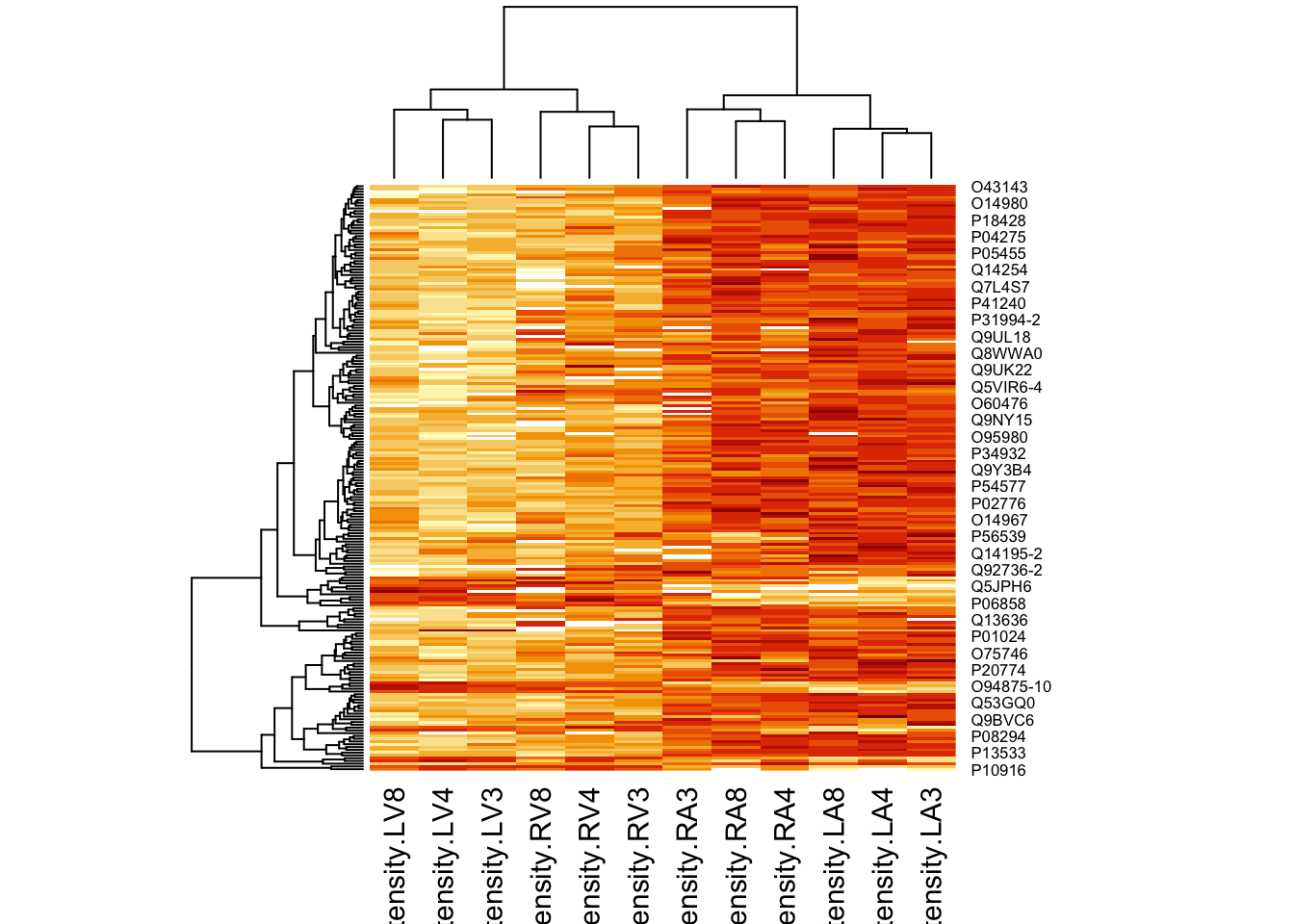
There are 211 proteins significantly differentially expressed at the 5% FDR level.
rowData(pe[["proteinRobust"]])$tissueV %>%
cbind(.,rowData(pe[["proteinRobust"]])$Protein.names) %>%
na.exclude %>%
filter(adjPval<0.05) %>%
arrange(pval) %>%
knitr::kable(.)| logFC | se | df | t | pval | adjPval | rowData(pe[[“proteinRobust”]])$Protein.names | |
|---|---|---|---|---|---|---|---|
| P08590 | 7.566945 | 0.4227679 | 9.117912 | 17.898582 | 0.0000000 | 0.0000417 | Myosin light chain 3 |
| P12883 | 4.333125 | 0.3637861 | 9.255194 | 11.911187 | 0.0000006 | 0.0006480 | Myosin-7 |
| P10916 | 6.908674 | 0.5190748 | 7.385413 | 13.309594 | 0.0000020 | 0.0013463 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| O75368 | -2.290376 | 0.2718512 | 9.251939 | -8.425109 | 0.0000123 | 0.0046575 | SH3 domain-binding glutamic acid-rich-like protein |
| P46821 | -2.182822 | 0.2654404 | 9.385413 | -8.223396 | 0.0000137 | 0.0046575 | Microtubule-associated protein 1B;MAP1B heavy chain;MAP1 light chain LC1 |
| Q6UWY5 | -3.253229 | 0.3888996 | 9.170135 | -8.365215 | 0.0000138 | 0.0046575 | Olfactomedin-like protein 1 |
| Q8N474 | -3.304647 | 0.3784419 | 7.847514 | -8.732243 | 0.0000261 | 0.0070102 | Secreted frizzled-related protein 1 |
| O95865 | -2.158932 | 0.2871753 | 9.385413 | -7.517818 | 0.0000288 | 0.0070102 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 |
| Q9ULL5-3 | -3.386308 | 0.3993822 | 7.893242 | -8.478865 | 0.0000311 | 0.0070102 | Proline-rich protein 12 |
| P02452 | -3.226193 | 0.4112622 | 8.367424 | -7.844614 | 0.0000392 | 0.0070838 | Collagen alpha-1(I) chain |
| O94875-10 | 2.321238 | 0.3122508 | 8.958355 | 7.433891 | 0.0000406 | 0.0070838 | Sorbin and SH3 domain-containing protein 2 |
| Q16647 | -2.787698 | 0.3824272 | 9.156725 | -7.289485 | 0.0000422 | 0.0070838 | Prostacyclin synthase |
| P29622 | -2.114870 | 0.2991501 | 9.385413 | -7.069595 | 0.0000474 | 0.0070838 | Kallistatin |
| P14854 | 2.300663 | 0.3037112 | 8.346841 | 7.575168 | 0.0000514 | 0.0070838 | Cytochrome c oxidase subunit 6B1 |
| P05546 | -1.915671 | 0.2708975 | 9.198639 | -7.071573 | 0.0000523 | 0.0070838 | Heparin cofactor 2 |
| P21810 | -3.380254 | 0.4548500 | 8.304055 | -7.431580 | 0.0000609 | 0.0077217 | Biglycan |
| P36955 | -2.323620 | 0.3325722 | 8.456893 | -6.986813 | 0.0000872 | 0.0097521 | Pigment epithelium-derived factor |
| P23083 | -4.274293 | 0.5581240 | 7.385413 | -7.658322 | 0.0000912 | 0.0097521 | Ig heavy chain V-I region V35 |
| P13533 | -4.313925 | 0.6516590 | 9.115028 | -6.619911 | 0.0000915 | 0.0097521 | Myosin-6 |
| Q15113 | -2.755021 | 0.4253583 | 9.188121 | -6.476942 | 0.0001043 | 0.0097521 | Procollagen C-endopeptidase enhancer 1 |
| Q9UBG0 | -2.576210 | 0.3963719 | 9.131671 | -6.499476 | 0.0001044 | 0.0097521 | C-type mannose receptor 2 |
| P00325 | -2.108814 | 0.3251595 | 9.141096 | -6.485476 | 0.0001057 | 0.0097521 | Alcohol dehydrogenase 1B |
| P07451 | -1.841539 | 0.2910138 | 9.385413 | -6.328012 | 0.0001136 | 0.0100264 | Carbonic anhydrase 3 |
| P51888 | -3.199549 | 0.4165978 | 6.959924 | -7.680186 | 0.0001217 | 0.0102934 | Prolargin |
| P24844 | -2.551801 | 0.4140400 | 9.330669 | -6.163176 | 0.0001427 | 0.0112127 | Myosin regulatory light polypeptide 9 |
| Q53GQ0 | -2.430853 | 0.3948274 | 9.334954 | -6.156748 | 0.0001436 | 0.0112127 | Very-long-chain 3-oxoacyl-CoA reductase |
| Q06828 | -4.209189 | 0.6775919 | 9.003303 | -6.211983 | 0.0001564 | 0.0116347 | Fibromodulin |
| Q8TBQ9 | -2.761806 | 0.4223033 | 8.170120 | -6.539863 | 0.0001643 | 0.0116347 | Protein kish-A |
| P51884 | -2.402234 | 0.3614104 | 7.936878 | -6.646831 | 0.0001672 | 0.0116347 | Lumican |
| P35442 | -2.334260 | 0.3655617 | 8.385413 | -6.385405 | 0.0001735 | 0.0116347 | Thrombospondin-2 |
| Q9UL18 | -2.602312 | 0.3982234 | 8.037138 | -6.534804 | 0.0001777 | 0.0116347 | Protein argonaute-1 |
| Q96LL9 | -2.426640 | 0.3958291 | 8.842792 | -6.130526 | 0.0001858 | 0.0117873 | DnaJ homolog subfamily C member 30 |
| P18428 | -2.002578 | 0.3365444 | 9.013977 | -5.950411 | 0.0002139 | 0.0131587 | Lipopolysaccharide-binding protein |
| P08294 | -2.756458 | 0.4335622 | 7.979491 | -6.357699 | 0.0002212 | 0.0132069 | Extracellular superoxide dismutase [Cu-Zn] |
| P36021 | -3.138301 | 0.5174695 | 8.385413 | -6.064707 | 0.0002493 | 0.0136858 | Monocarboxylate transporter 8 |
| P02743 | -2.273778 | 0.3999916 | 9.385413 | -5.684565 | 0.0002568 | 0.0136858 | Serum amyloid P-component;Serum amyloid P-component(1-203) |
| Q8WWA0 | -5.806975 | 0.9532620 | 8.244689 | -6.091688 | 0.0002588 | 0.0136858 | Intelectin-1 |
| Q92736-2 | -3.330740 | 0.5744993 | 8.908498 | -5.797640 | 0.0002706 | 0.0136858 | Ryanodine receptor 2 |
| O60760 | -3.500029 | 0.5842390 | 8.385413 | -5.990748 | 0.0002715 | 0.0136858 | Hematopoietic prostaglandin D synthase |
| Q9UGT4 | -2.283615 | 0.4020386 | 9.193722 | -5.680088 | 0.0002789 | 0.0136858 | Sushi domain-containing protein 2 |
| Q14764 | -1.625976 | 0.2901417 | 9.385413 | -5.604074 | 0.0002855 | 0.0136858 | Major vault protein |
| O95980 | -2.552421 | 0.4176977 | 7.980660 | -6.110688 | 0.0002889 | 0.0136858 | Reversion-inducing cysteine-rich protein with Kazal motifs |
| Q92508 | 4.071001 | 0.5712249 | 6.385413 | 7.126791 | 0.0002899 | 0.0136858 | Piezo-type mechanosensitive ion channel component 1 |
| Q9BW30 | -2.738684 | 0.4828316 | 9.049210 | -5.672131 | 0.0002988 | 0.0137863 | Tubulin polymerization-promoting protein family member 3 |
| P12110 | -2.136456 | 0.3667316 | 8.535409 | -5.825667 | 0.0003078 | 0.0138861 | Collagen alpha-2(VI) chain |
| P40261 | -2.306727 | 0.4169709 | 9.340024 | -5.532105 | 0.0003196 | 0.0141028 | Nicotinamide N-methyltransferase |
| P05997 | -3.068864 | 0.5429778 | 8.864434 | -5.651913 | 0.0003308 | 0.0141028 | Collagen alpha-2(V) chain |
| Q9P2B2 | -2.047509 | 0.3731311 | 9.385413 | -5.487374 | 0.0003335 | 0.0141028 | Prostaglandin F2 receptor negative regulator |
| Q9NZ01 | -2.346040 | 0.4161143 | 8.691035 | -5.637970 | 0.0003618 | 0.0148853 | Very-long-chain enoyl-CoA reductase |
| P46060 | -1.970642 | 0.3436046 | 8.385413 | -5.735204 | 0.0003666 | 0.0148853 | Ran GTPase-activating protein 1 |
| P31994-2 | -1.655566 | 0.3017732 | 8.992524 | -5.486128 | 0.0003882 | 0.0152351 | Low affinity immunoglobulin gamma Fc region receptor II-b |
| O14967 | -2.393131 | 0.4223134 | 8.430438 | -5.666718 | 0.0003903 | 0.0152351 | Calmegin |
| P00748 | -2.003056 | 0.3702529 | 9.091376 | -5.409968 | 0.0004128 | 0.0156430 | Coagulation factor XII;Coagulation factor XIIa heavy chain;Beta-factor XIIa part 1;Coagulation factor XIIa light chain |
| Q07954 | -1.674832 | 0.3146216 | 9.385413 | -5.323322 | 0.0004161 | 0.0156430 | Prolow-density lipoprotein receptor-related protein 1;Low-density lipoprotein receptor-related protein 1 85 kDa subunit;Low-density lipoprotein receptor-related protein 1 515 kDa subunit;Low-density lipoprotein receptor-related protein 1 intracellular domain |
| Q9UBB5 | 2.673301 | 0.4015137 | 6.385413 | 6.658059 | 0.0004277 | 0.0157858 | Methyl-CpG-binding domain protein 2 |
| O43677 | -2.758090 | 0.4792076 | 7.933870 | -5.755523 | 0.0004396 | 0.0159350 | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial |
| Q92604 | -2.270281 | 0.4316583 | 9.345516 | -5.259442 | 0.0004605 | 0.0163992 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 |
| Q8NAT1 | -1.416451 | 0.2751694 | 9.385413 | -5.147559 | 0.0005298 | 0.0181178 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 |
| O00264 | -1.999242 | 0.3827164 | 9.079390 | -5.223821 | 0.0005311 | 0.0181178 | Membrane-associated progesterone receptor component 1 |
| Q92621 | -1.755581 | 0.3415656 | 9.385413 | -5.139808 | 0.0005355 | 0.0181178 | Nuclear pore complex protein Nup205 |
| Q8WZA9 | -1.502201 | 0.2907398 | 9.101344 | -5.166822 | 0.0005690 | 0.0186760 | Immunity-related GTPase family Q protein |
| Q9BXN1 | -2.670923 | 0.5132981 | 8.937487 | -5.203455 | 0.0005742 | 0.0186760 | Asporin |
| Q14195-2 | -2.527278 | 0.4943182 | 9.262403 | -5.112654 | 0.0005796 | 0.0186760 | Dihydropyrimidinase-related protein 3 |
| O00180 | -4.096722 | 0.7724957 | 8.385413 | -5.303229 | 0.0006215 | 0.0197142 | Potassium channel subfamily K member 1 |
| Q8TBP6 | -1.893243 | 0.3673308 | 8.851865 | -5.154053 | 0.0006323 | 0.0197464 | Solute carrier family 25 member 40 |
| Q9GZY4 | -3.167768 | 0.5759744 | 7.703311 | -5.499842 | 0.0006539 | 0.0198708 | Cytochrome c oxidase assembly factor 1 homolog |
| P41240 | -1.569148 | 0.3075870 | 8.922337 | -5.101476 | 0.0006615 | 0.0198708 | Tyrosine-protein kinase CSK |
| Q9NY15 | -2.177423 | 0.4377625 | 9.336638 | -4.973981 | 0.0006860 | 0.0198708 | Stabilin-1 |
| Q96C86 | -1.515572 | 0.3020943 | 9.145534 | -5.016883 | 0.0006878 | 0.0198708 | m7GpppX diphosphatase |
| P02747 | -2.680914 | 0.4712559 | 7.138784 | -5.688871 | 0.0006937 | 0.0198708 | Complement C1q subcomponent subunit C |
| P06858 | 1.795244 | 0.3596173 | 9.218266 | 4.992094 | 0.0006950 | 0.0198708 | Lipoprotein lipase |
| O43464 | -1.900721 | 0.3799226 | 9.072346 | -5.002915 | 0.0007184 | 0.0201585 | Serine protease HTRA2, mitochondrial |
| O14980 | -1.230352 | 0.2498369 | 9.382448 | -4.924620 | 0.0007249 | 0.0201585 | Exportin-1 |
| Q8WY22 | -1.839442 | 0.3608464 | 8.385413 | -5.097577 | 0.0008064 | 0.0221211 | BRI3-binding protein |
| P04083 | -1.524538 | 0.3158141 | 9.385413 | -4.827329 | 0.0008320 | 0.0225191 | Annexin A1 |
| P48681 | -1.330081 | 0.2756444 | 9.321612 | -4.825351 | 0.0008507 | 0.0227223 | Nestin |
| P02775 | -1.952126 | 0.4075050 | 9.385413 | -4.790435 | 0.0008772 | 0.0229385 | Platelet basic protein;Connective tissue-activating peptide III;TC-2;Connective tissue-activating peptide III(1-81);Beta-thromboglobulin;Neutrophil-activating peptide 2(74);Neutrophil-activating peptide 2(73);Neutrophil-activating peptide 2;TC-1;Neutrophil-activating peptide 2(1-66);Neutrophil-activating peptide 2(1-63) |
| Q7L4S7 | -1.747450 | 0.3202677 | 7.157555 | -5.456217 | 0.0008814 | 0.0229385 | Protein ARMCX6 |
| P46063 | -1.386912 | 0.2873890 | 9.154885 | -4.825907 | 0.0008948 | 0.0229922 | ATP-dependent DNA helicase Q1 |
| A6NMZ7 | -2.267329 | 0.4773591 | 9.385413 | -4.749734 | 0.0009302 | 0.0232949 | Collagen alpha-6(VI) chain |
| Q5NDL2 | -2.337629 | 0.4766026 | 8.686198 | -4.904777 | 0.0009340 | 0.0232949 | EGF domain-specific O-linked N-acetylglucosamine transferase |
| P56539 | -2.051918 | 0.4159611 | 8.553679 | -4.932956 | 0.0009410 | 0.0232949 | Caveolin-3 |
| Q9UNW9 | 4.097053 | 0.8545595 | 9.054127 | 4.794344 | 0.0009650 | 0.0236026 | RNA-binding protein Nova-2 |
| P01699 | -4.025961 | 0.6965009 | 6.314916 | -5.780266 | 0.0009792 | 0.0236632 | Ig lambda chain V-I region VOR |
| Q9HAV4 | -2.205274 | 0.4388018 | 7.987893 | -5.025671 | 0.0010243 | 0.0244630 | Exportin-5 |
| Q92681 | -2.153054 | 0.4353071 | 8.133279 | -4.946057 | 0.0010730 | 0.0253280 | Regulatory solute carrier protein family 1 member 1 |
| P49207 | -1.534829 | 0.3249990 | 8.970773 | -4.722565 | 0.0010949 | 0.0255013 | 60S ribosomal protein L34 |
| P50552 | -1.523119 | 0.3245142 | 9.074476 | -4.693535 | 0.0011055 | 0.0255013 | Vasodilator-stimulated phosphoprotein |
| Q5VIR6-4 | -1.755057 | 0.3758054 | 9.130015 | -4.670123 | 0.0011243 | 0.0256432 | Vacuolar protein sorting-associated protein 53 homolog |
| Q96H79 | -3.021670 | 0.5420367 | 6.334231 | -5.574659 | 0.0011785 | 0.0265817 | Zinc finger CCCH-type antiviral protein 1-like |
| O75828 | -1.641739 | 0.3592450 | 9.385413 | -4.569971 | 0.0012087 | 0.0269634 | Carbonyl reductase [NADPH] 3 |
| O15230 | -1.415011 | 0.3058820 | 9.044210 | -4.626004 | 0.0012277 | 0.0270900 | Laminin subunit alpha-5 |
| Q5M9N0 | -3.537066 | 0.7427497 | 8.350040 | -4.762123 | 0.0012639 | 0.0275884 | Coiled-coil domain-containing protein 158 |
| P04196 | -1.917181 | 0.3881943 | 7.683140 | -4.938714 | 0.0012797 | 0.0276358 | Histidine-rich glycoprotein |
| Q53GG5-2 | -2.923374 | 0.6214767 | 8.429675 | -4.703916 | 0.0013308 | 0.0282214 | PDZ and LIM domain protein 3 |
| Q6SZW1 | -2.563687 | 0.5126523 | 7.385413 | -5.000831 | 0.0013346 | 0.0282214 | Sterile alpha and TIR motif-containing protein 1 |
| Q15274 | -1.928288 | 0.3958755 | 7.757883 | -4.870945 | 0.0013525 | 0.0283041 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] |
| Q9BXR6 | -2.352662 | 0.5252419 | 9.382448 | -4.479196 | 0.0013831 | 0.0283325 | Complement factor H-related protein 5 |
| Q9ULC3 | -1.674808 | 0.3626400 | 8.669265 | -4.618376 | 0.0013874 | 0.0283325 | Ras-related protein Rab-23 |
| Q9Y5U8 | -3.988833 | 0.8897580 | 9.327097 | -4.483053 | 0.0013957 | 0.0283325 | Mitochondrial pyruvate carrier 1 |
| P34932 | -1.163429 | 0.2608932 | 9.385413 | -4.459407 | 0.0014232 | 0.0286058 | Heat shock 70 kDa protein 4 |
| Q9BUF5 | -1.934378 | 0.4345654 | 9.217423 | -4.451293 | 0.0015060 | 0.0298342 | Tubulin beta-6 chain |
| Q8TDB6 | -1.611315 | 0.3613218 | 9.153547 | -4.459500 | 0.0015138 | 0.0298342 | E3 ubiquitin-protein ligase DTX3L |
| P45877 | -1.259792 | 0.2861984 | 9.343571 | -4.401814 | 0.0015676 | 0.0305974 | Peptidyl-prolyl cis-trans isomerase C |
| P04003 | -1.479053 | 0.3338506 | 9.090898 | -4.430283 | 0.0016066 | 0.0310601 | C4b-binding protein alpha chain |
| P06727 | -1.358368 | 0.3048521 | 8.884167 | -4.455826 | 0.0016384 | 0.0312643 | Apolipoprotein A-IV |
| P09619 | -1.675728 | 0.3807310 | 9.154490 | -4.401345 | 0.0016479 | 0.0312643 | Platelet-derived growth factor receptor beta |
| Q8N5M1 | -2.022143 | 0.4573254 | 8.937206 | -4.421672 | 0.0016958 | 0.0316965 | ATP synthase mitochondrial F1 complex assembly factor 2 |
| Q6YN16 | 1.527167 | 0.3496754 | 9.222955 | 4.367385 | 0.0017019 | 0.0316965 | Hydroxysteroid dehydrogenase-like protein 2 |
| P01042 | -2.256316 | 0.4659486 | 7.210294 | -4.842414 | 0.0017231 | 0.0317986 | Kininogen-1;Kininogen-1 heavy chain;T-kinin;Bradykinin;Lysyl-bradykinin;Kininogen-1 light chain;Low molecular weight growth-promoting factor |
| P08582 | -1.663860 | 0.3857501 | 9.299314 | -4.313310 | 0.0018093 | 0.0329078 | Melanotransferrin |
| P30405 | -1.540274 | 0.3587630 | 9.385413 | -4.293290 | 0.0018252 | 0.0329078 | Peptidyl-prolyl cis-trans isomerase F, mitochondrial |
| P14550 | -1.394080 | 0.3100035 | 8.301450 | -4.496981 | 0.0018318 | 0.0329078 | Alcohol dehydrogenase [NADP(+)] |
| Q9UQ35 | -1.367511 | 0.3203946 | 9.385413 | -4.268208 | 0.0018958 | 0.0334035 | Serine/arginine repetitive matrix protein 2 |
| P36551 | -1.338543 | 0.3106105 | 9.112223 | -4.309393 | 0.0019078 | 0.0334035 | Oxygen-dependent coproporphyrinogen-III oxidase, mitochondrial |
| P14555 | -4.594903 | 0.9455710 | 6.867951 | -4.859396 | 0.0019377 | 0.0334035 | Phospholipase A2, membrane associated |
| Q08945 | -2.071352 | 0.4670698 | 8.385413 | -4.434781 | 0.0019453 | 0.0334035 | FACT complex subunit SSRP1 |
| P24311 | 1.725765 | 0.4048607 | 9.295713 | 4.262615 | 0.0019540 | 0.0334035 | Cytochrome c oxidase subunit 7B, mitochondrial |
| P02461 | -3.597666 | 0.8096938 | 8.323445 | -4.443242 | 0.0019581 | 0.0334035 | Collagen alpha-1(III) chain |
| Q9H1E5 | -1.388829 | 0.3280259 | 9.385413 | -4.233902 | 0.0019969 | 0.0337811 | Thioredoxin-related transmembrane protein 4 |
| O00303 | -1.477605 | 0.3444231 | 9.008150 | -4.290088 | 0.0020156 | 0.0338155 | Eukaryotic translation initiation factor 3 subunit F |
| Q9Y6X5 | -1.411435 | 0.3280595 | 8.750753 | -4.302375 | 0.0021171 | 0.0348421 | Bis(5-adenosyl)-triphosphatase ENPP4 |
| P15924 | 1.274221 | 0.3034579 | 9.350459 | 4.199004 | 0.0021231 | 0.0348421 | Desmoplakin |
| O75746 | -1.578482 | 0.3695373 | 8.893946 | -4.271509 | 0.0021327 | 0.0348421 | Calcium-binding mitochondrial carrier protein Aralar1 |
| P05455 | -1.204229 | 0.2854061 | 9.178555 | -4.219353 | 0.0021455 | 0.0348421 | Lupus La protein |
| Q2TAA5 | -1.319245 | 0.3143146 | 9.249693 | -4.197212 | 0.0021804 | 0.0349673 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase |
| P04004 | -1.633973 | 0.3854552 | 8.981078 | -4.239073 | 0.0021876 | 0.0349673 | Vitronectin;Vitronectin V65 subunit;Vitronectin V10 subunit;Somatomedin-B |
| P01031 | -1.322378 | 0.3017394 | 8.183937 | -4.382518 | 0.0022165 | 0.0351530 | Complement C5;Complement C5 beta chain;Complement C5 alpha chain;C5a anaphylatoxin;Complement C5 alpha chain |
| Q5JPH6 | 3.506017 | 0.7033283 | 6.212234 | 4.984894 | 0.0022530 | 0.0354450 | Probable glutamate–tRNA ligase, mitochondrial |
| P49458 | -2.320823 | 0.5006356 | 7.120983 | -4.635753 | 0.0022768 | 0.0354450 | Signal recognition particle 9 kDa protein |
| P04209 | 1.316898 | 0.3151619 | 9.167154 | 4.178483 | 0.0022873 | 0.0354450 | Ig lambda chain V-II region NIG-84 |
| Q04721 | 1.531039 | 0.3675191 | 9.201253 | 4.165877 | 0.0023123 | 0.0355608 | Neurogenic locus notch homolog protein 2;Notch 2 extracellular truncation;Notch 2 intracellular domain |
| Q9UKS6 | 1.443688 | 0.3439564 | 8.943424 | 4.197299 | 0.0023485 | 0.0358449 | Protein kinase C and casein kinase substrate in neurons protein 3 |
| Q9BTV4 | -1.810463 | 0.4381260 | 9.284468 | -4.132288 | 0.0023862 | 0.0361484 | Transmembrane protein 43 |
| O75348 | -1.846417 | 0.4493725 | 9.377764 | -4.108879 | 0.0024210 | 0.0364041 | V-type proton ATPase subunit G 1 |
| Q12996 | -1.667403 | 0.3663886 | 7.207709 | -4.550913 | 0.0024442 | 0.0364833 | Cleavage stimulation factor subunit 3 |
| P60468 | -1.522924 | 0.3550773 | 8.241407 | -4.288994 | 0.0024787 | 0.0367280 | Protein transport protein Sec61 subunit beta |
| Q6ZSY5 | -1.520789 | 0.3699133 | 9.170791 | -4.111204 | 0.0025292 | 0.0370388 | Protein phosphatase 1 regulatory subunit 3F |
| P62760 | -1.929645 | 0.4686939 | 9.120970 | -4.117068 | 0.0025362 | 0.0370388 | Visinin-like protein 1 |
| Q14019 | -3.973669 | 0.8363384 | 6.385413 | -4.751270 | 0.0026690 | 0.0387005 | Coactosin-like protein |
| Q13478 | -1.620268 | 0.3970937 | 8.994941 | -4.080316 | 0.0027600 | 0.0397356 | Interleukin-18 receptor 1 |
| P01034 | -1.689330 | 0.4126063 | 8.837162 | -4.094289 | 0.0028058 | 0.0401104 | Cystatin-C |
| Q53T59 | -1.852124 | 0.4344248 | 7.876061 | -4.263395 | 0.0028487 | 0.0402275 | HCLS1-binding protein 3 |
| P02776 | -1.748717 | 0.4069507 | 7.718405 | -4.297123 | 0.0028536 | 0.0402275 | Platelet factor 4;Platelet factor 4, short form |
| P26447 | -1.877769 | 0.4593883 | 8.774968 | -4.087542 | 0.0028764 | 0.0402690 | Protein S100-A4 |
| Q9Y2Z0 | -1.757978 | 0.4412870 | 9.385413 | -3.983751 | 0.0029317 | 0.0405660 | Suppressor of G2 allele of SKP1 homolog |
| O75475 | -1.951060 | 0.4885410 | 9.296501 | -3.993647 | 0.0029426 | 0.0405660 | PC4 and SFRS1-interacting protein |
| Q1KMD3 | -1.494563 | 0.3712572 | 9.051770 | -4.025682 | 0.0029575 | 0.0405660 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 |
| P01024 | -1.148297 | 0.2873908 | 9.197920 | -3.995596 | 0.0029972 | 0.0408343 | Complement C3;Complement C3 beta chain;C3-beta-c;Complement C3 alpha chain;C3a anaphylatoxin;Acylation stimulating protein;Complement C3b alpha chain;Complement C3c alpha chain fragment 1;Complement C3dg fragment;Complement C3g fragment;Complement C3d fragment;Complement C3f fragment;Complement C3c alpha chain fragment 2 |
| P54577 | -1.309794 | 0.3255176 | 8.964139 | -4.023729 | 0.0030257 | 0.0409480 | Tyrosine–tRNA ligase, cytoplasmic;Tyrosine–tRNA ligase, cytoplasmic, N-terminally processed |
| P54652 | -1.585922 | 0.3969039 | 9.007166 | -3.995732 | 0.0031255 | 0.0420185 | Heat shock-related 70 kDa protein 2 |
| P25311 | -1.476798 | 0.3658030 | 8.673237 | -4.037141 | 0.0031737 | 0.0422552 | Zinc-alpha-2-glycoprotein |
| Q9UK22 | -1.968951 | 0.4285590 | 6.372396 | -4.594351 | 0.0031847 | 0.0422552 | F-box only protein 2 |
| O94919 | -1.158582 | 0.2941090 | 9.248102 | -3.939294 | 0.0032331 | 0.0425105 | Endonuclease domain-containing 1 protein |
| P03950 | -2.214295 | 0.5408135 | 8.216113 | -4.094379 | 0.0032737 | 0.0425105 | Angiogenin |
| O15111 | -1.770061 | 0.4097076 | 7.141811 | -4.320302 | 0.0033195 | 0.0425105 | Inhibitor of nuclear factor kappa-B kinase subunit alpha |
| P07357 | -1.154912 | 0.2950375 | 9.290174 | -3.914458 | 0.0033303 | 0.0425105 | Complement component C8 alpha chain |
| O15116 | -3.262825 | 0.6533382 | 5.390877 | -4.994083 | 0.0033442 | 0.0425105 | U6 snRNA-associated Sm-like protein LSm1 |
| Q13641 | -2.094962 | 0.4829370 | 7.050947 | -4.337963 | 0.0033469 | 0.0425105 | Trophoblast glycoprotein |
| Q9BS26 | -1.226358 | 0.3132596 | 9.249914 | -3.914830 | 0.0033564 | 0.0425105 | Endoplasmic reticulum resident protein 44 |
| Q14011 | -2.439155 | 0.5991974 | 8.231467 | -4.070703 | 0.0033715 | 0.0425105 | Cold-inducible RNA-binding protein |
| P12814 | -1.726195 | 0.4239901 | 8.144626 | -4.071308 | 0.0034446 | 0.0431638 | Alpha-actinin-1 |
| P25940 | -1.680300 | 0.4292504 | 9.043783 | -3.914499 | 0.0035074 | 0.0436810 | Collagen alpha-3(V) chain |
| Q9UBI9 | -2.822134 | 0.6797511 | 7.613412 | -4.151717 | 0.0035670 | 0.0441523 | Headcase protein homolog |
| Q96PK6 | -1.284104 | 0.3320346 | 9.148490 | -3.867380 | 0.0036883 | 0.0448514 | RNA-binding protein 14 |
| Q7LBR1 | -2.851457 | 0.7432377 | 9.385413 | -3.836534 | 0.0036895 | 0.0448514 | Charged multivesicular body protein 1b |
| P07384 | -1.106938 | 0.2881747 | 9.333139 | -3.841205 | 0.0037006 | 0.0448514 | Calpain-1 catalytic subunit |
| O60262 | -1.514780 | 0.3809638 | 8.388576 | -3.976177 | 0.0037118 | 0.0448514 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-7 |
| Q86VU5 | 1.574090 | 0.4112246 | 9.385413 | 3.827810 | 0.0037405 | 0.0449299 | Catechol O-methyltransferase domain-containing protein 1 |
| Q13636 | -3.085924 | 0.6708563 | 5.941397 | -4.599978 | 0.0037856 | 0.0452041 | Ras-related protein Rab-31 |
| O43143 | -1.010978 | 0.2650086 | 9.289616 | -3.814887 | 0.0038896 | 0.0458869 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 |
| P20774 | -2.236773 | 0.5631895 | 8.210882 | -3.971618 | 0.0039003 | 0.0458869 | Mimecan |
| P08311 | -1.326822 | 0.3393339 | 8.579011 | -3.910077 | 0.0039106 | 0.0458869 | Cathepsin G |
| A8MTT3 | -3.368826 | 0.8032193 | 7.070471 | -4.194155 | 0.0039754 | 0.0461448 | Protein CEBPZOS |
| Q9HB40 | -2.173414 | 0.4870876 | 6.179009 | -4.462060 | 0.0039780 | 0.0461448 | Retinoid-inducible serine carboxypeptidase |
| O60476 | -2.858033 | 0.6987082 | 7.449546 | -4.090453 | 0.0040536 | 0.0462919 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB |
| Q96CS3 | -1.329185 | 0.3488970 | 9.119924 | -3.809678 | 0.0040557 | 0.0462919 | FAS-associated factor 2 |
| Q04941 | -1.954543 | 0.5132443 | 9.127049 | -3.808212 | 0.0040591 | 0.0462919 | Proteolipid protein 2 |
| Q8NFQ8 | -1.244891 | 0.3112852 | 7.843418 | -3.999198 | 0.0041168 | 0.0466119 | Torsin-1A-interacting protein 2 |
| Q9Y3B4 | -1.254563 | 0.3315970 | 9.218746 | -3.783396 | 0.0041429 | 0.0466119 | Splicing factor 3B subunit 6 |
| O00567 | -2.329642 | 0.6192522 | 9.376799 | -3.762024 | 0.0041560 | 0.0466119 | Nucleolar protein 56 |
| Q9UKX3 | 2.040096 | 0.5408257 | 9.208912 | 3.772187 | 0.0042241 | 0.0471152 | Myosin-13 |
| P56199 | -1.282712 | 0.3337866 | 8.647866 | -3.842911 | 0.0042549 | 0.0471986 | Integrin alpha-1 |
| P14543 | -1.384551 | 0.3678338 | 9.204669 | -3.764065 | 0.0042816 | 0.0472368 | Nidogen-1 |
| Q9Y2D4 | -1.989829 | 0.5205860 | 8.653139 | -3.822287 | 0.0043830 | 0.0479403 | Exocyst complex component 6B |
| Q6P1N0 | -1.699604 | 0.4098304 | 6.936107 | -4.147091 | 0.0043979 | 0.0479403 | Coiled-coil and C2 domain-containing protein 1A |
| Q8NBF2 | -1.164826 | 0.3132700 | 9.379760 | -3.718280 | 0.0044515 | 0.0479403 | NHL repeat-containing protein 2 |
| P62745 | -1.495470 | 0.3945560 | 8.788016 | -3.790261 | 0.0044705 | 0.0479403 | Rho-related GTP-binding protein RhoB |
| Q14254 | -1.392125 | 0.3457572 | 7.385413 | -4.026309 | 0.0044916 | 0.0479403 | Flotillin-2 |
| Q9Y287 | -1.918702 | 0.5175377 | 9.385413 | -3.707366 | 0.0045246 | 0.0479403 | Integral membrane protein 2B;BRI2, membrane form;BRI2 intracellular domain;BRI2C, soluble form;Bri23 peptide |
| P49773 | -1.295274 | 0.3495513 | 9.385413 | -3.705532 | 0.0045377 | 0.0479403 | Histidine triad nucleotide-binding protein 1 |
| Q9BVC6 | -1.637083 | 0.4433031 | 9.341936 | -3.692921 | 0.0046665 | 0.0479403 | Transmembrane protein 109 |
| P08670 | -1.482725 | 0.3924566 | 8.643328 | -3.778060 | 0.0046928 | 0.0479403 | Vimentin |
| P50991 | -1.786149 | 0.4735903 | 8.685140 | -3.771507 | 0.0046981 | 0.0479403 | T-complex protein 1 subunit delta |
| P23434 | 1.126822 | 0.3062675 | 9.385413 | 3.679208 | 0.0047316 | 0.0479403 | Glycine cleavage system H protein, mitochondrial |
| Q6IC98 | -1.126652 | 0.3015080 | 8.888308 | -3.736724 | 0.0047525 | 0.0479403 | GRAM domain-containing protein 4 |
| Q8IXW5 | 3.388152 | 0.8332024 | 7.009655 | 4.066422 | 0.0047569 | 0.0479403 | Putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 |
| Q6P587 | 5.677610 | 1.3549193 | 6.518479 | 4.190368 | 0.0047840 | 0.0479403 | Acylpyruvase FAHD1, mitochondrial |
| P01008 | -1.547192 | 0.4205447 | 9.310907 | -3.679019 | 0.0047975 | 0.0479403 | Antithrombin-III |
| P04843 | -2.051639 | 0.5456574 | 8.660976 | -3.759939 | 0.0048046 | 0.0479403 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 |
| Q96FJ2 | 1.591193 | 0.4148520 | 8.139895 | 3.835568 | 0.0048186 | 0.0479403 | Dynein light chain 2, cytoplasmic |
| Q7Z3T8 | -1.576843 | 0.4157303 | 8.413206 | -3.792946 | 0.0048217 | 0.0479403 | Zinc finger FYVE domain-containing protein 16 |
| P51665 | -1.274787 | 0.3456226 | 9.194991 | -3.688379 | 0.0048292 | 0.0479403 | 26S proteasome non-ATPase regulatory subunit 7 |
| P80723 | -1.638540 | 0.4231920 | 7.910668 | -3.871858 | 0.0048319 | 0.0479403 | Brain acid soluble protein 1 |
| O95486 | -1.500300 | 0.4068449 | 9.187800 | -3.687646 | 0.0048413 | 0.0479403 | Protein transport protein Sec24A |
| Q14258 | -1.359986 | 0.3561261 | 8.157903 | -3.818833 | 0.0049155 | 0.0483626 | E3 ubiquitin/ISG15 ligase TRIM25 |
| P04275 | -1.054100 | 0.2849243 | 8.991784 | -3.699577 | 0.0049316 | 0.0483626 | von Willebrand factor;von Willebrand antigen 2 |
| Q16082 | -1.361982 | 0.3713096 | 9.207589 | -3.668050 | 0.0049745 | 0.0485490 | Heat shock protein beta-2 |
| P32119 | -1.348354 | 0.3464470 | 7.636769 | -3.891950 | 0.0050302 | 0.0488581 | Peroxiredoxin-2 |
| P05060 | -2.649749 | 0.6720512 | 7.305570 | -3.942778 | 0.0051268 | 0.0495588 | Secretogranin-1;PE-11;GAWK peptide;CCB peptide |
| Q9HCB6 | -1.808880 | 0.4871150 | 8.648303 | -3.713455 | 0.0051666 | 0.0497073 | Spondin-1 |
4.4 Evaluate results contrast \(\log_2 FC_{V-A}^R\)
4.4.1 Volcano-plot
volcanoRight <- ggplot(rowData(pe[["proteinRobust"]])$"tissueV + locationR:tissueV",
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) + theme_minimal()
volcanoRight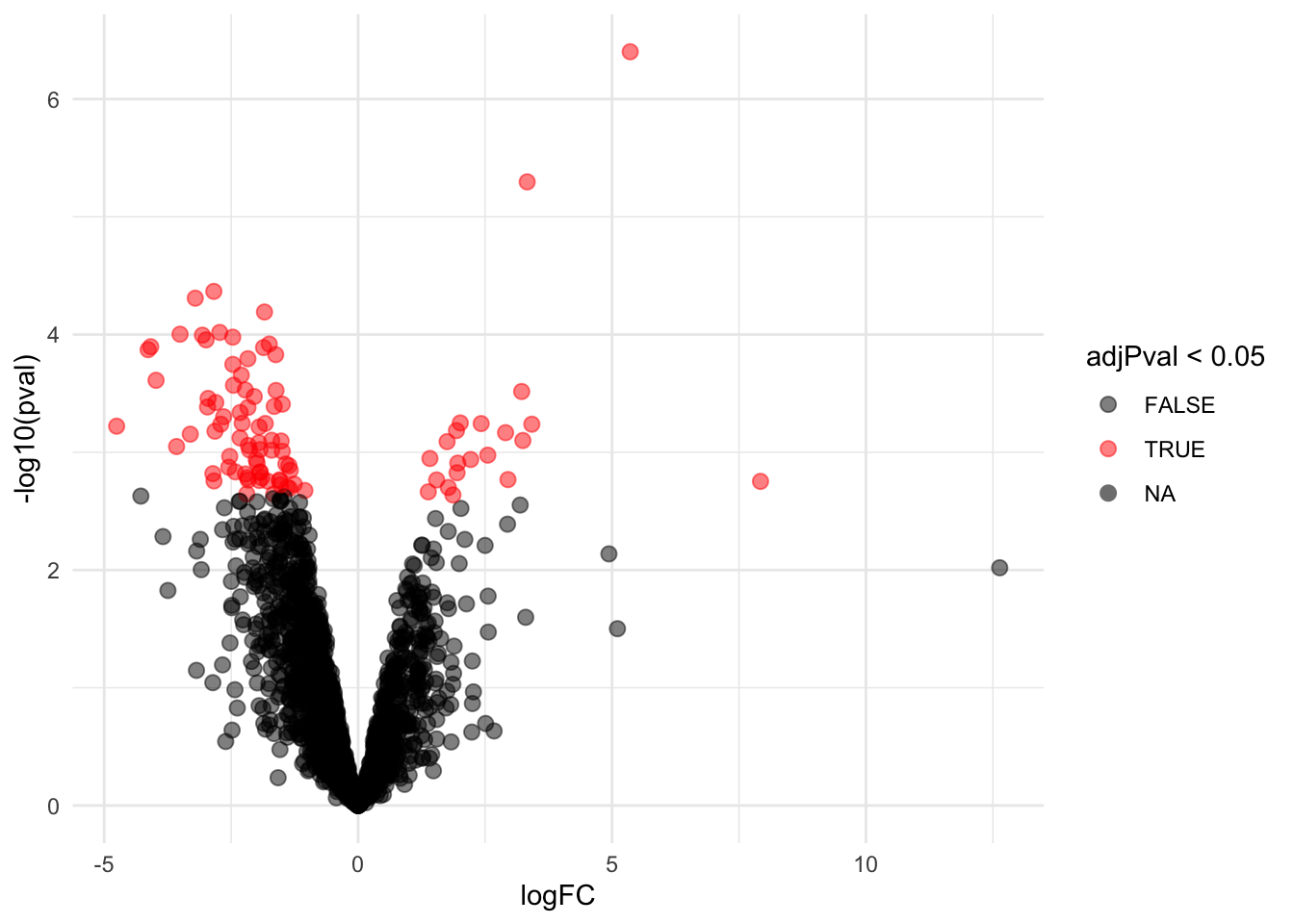
4.4.2 Heatmap
We first select the names of the proteins that were declared signficant.
sigNamesRight <- rowData(pe[["proteinRobust"]])$"tissueV + locationR:tissueV" %>%
rownames_to_column("proteinRobust") %>%
filter(adjPval<0.05) %>%
pull(proteinRobust)
heatmap(assay(pe[["proteinRobust"]])[sigNamesRight, ])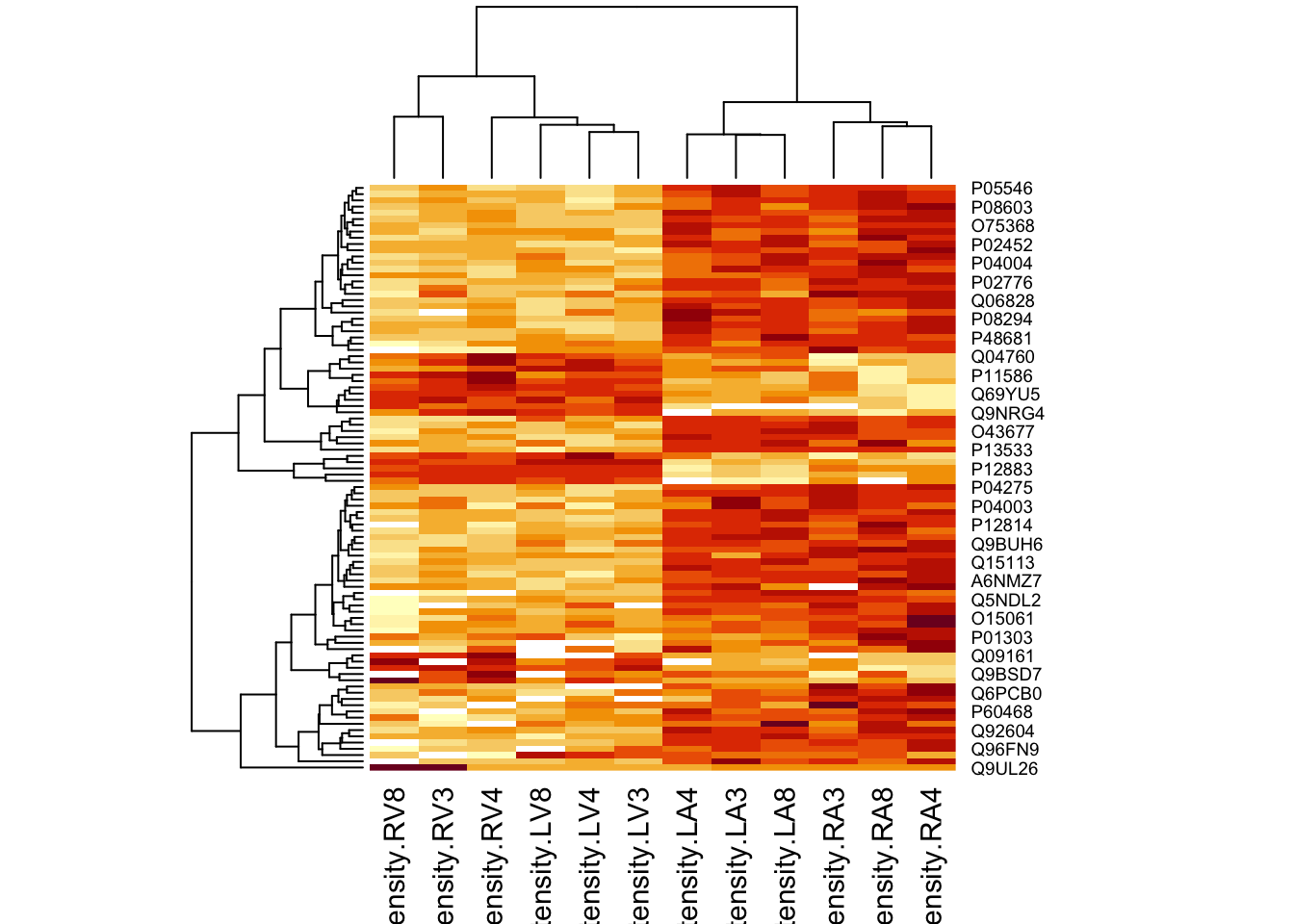
There are 94 proteins significantly differentially expressed at the 5% FDR level.
rowData(pe[["proteinRobust"]])$"tissueV + locationR:tissueV" %>%
cbind(.,rowData(pe[["proteinRobust"]])$Protein.names) %>%
na.exclude %>%
filter(adjPval<0.05) %>%
arrange(pval) %>%
knitr::kable(.)| logFC | se | df | t | pval | adjPval | rowData(pe[[“proteinRobust”]])$Protein.names | |
|---|---|---|---|---|---|---|---|
| P08590 | 5.358077 | 0.4191438 | 9.117912 | 12.783387 | 0.0000004 | 0.0008046 | Myosin light chain 3 |
| P06858 | 3.329246 | 0.3542730 | 9.218266 | 9.397404 | 0.0000051 | 0.0051277 | Lipoprotein lipase |
| P02776 | -2.841711 | 0.3443690 | 7.718405 | -8.251937 | 0.0000430 | 0.0194907 | Platelet factor 4;Platelet factor 4, short form |
| P35442 | -3.207091 | 0.4221143 | 8.385413 | -7.597683 | 0.0000491 | 0.0194907 | Thrombospondin-2 |
| O75368 | -1.844075 | 0.2686680 | 9.251939 | -6.863771 | 0.0000643 | 0.0194907 | SH3 domain-binding glutamic acid-rich-like protein |
| P54652 | -2.725246 | 0.4112417 | 9.007166 | -6.626871 | 0.0000959 | 0.0194907 | Heat shock-related 70 kDa protein 2 |
| Q9ULD0 | -3.507028 | 0.3938799 | 6.128068 | -8.903799 | 0.0000993 | 0.0194907 | 2-oxoglutarate dehydrogenase-like, mitochondrial |
| A6NMZ7 | -3.065958 | 0.4773591 | 9.385413 | -6.422751 | 0.0001012 | 0.0194907 | Collagen alpha-6(VI) chain |
| Q6UWY5 | -2.469667 | 0.3813094 | 9.170135 | -6.476806 | 0.0001053 | 0.0194907 | Olfactomedin-like protein 1 |
| P21810 | -2.994207 | 0.4377152 | 8.304055 | -6.840536 | 0.0001110 | 0.0194907 | Biglycan |
| P05546 | -1.750184 | 0.2755089 | 9.198639 | -6.352548 | 0.0001204 | 0.0194907 | Heparin cofactor 2 |
| P13533 | -4.083951 | 0.6439331 | 9.115028 | -6.342197 | 0.0001269 | 0.0194907 | Myosin-6 |
| P29622 | -1.862155 | 0.2991501 | 9.385413 | -6.224819 | 0.0001290 | 0.0194907 | Kallistatin |
| Q06828 | -4.135476 | 0.6523980 | 9.003303 | -6.338886 | 0.0001344 | 0.0194907 | Fibromodulin |
| P46821 | -1.622963 | 0.2654404 | 9.385413 | -6.114227 | 0.0001481 | 0.0200375 | Microtubule-associated protein 1B;MAP1B heavy chain;MAP1 light chain LC1 |
| P28066 | -2.170776 | 0.3390487 | 8.500773 | -6.402548 | 0.0001604 | 0.0203510 | Proteasome subunit alpha type-5 |
| Q9UGT4 | -2.466535 | 0.4090800 | 9.193722 | -6.029468 | 0.0001792 | 0.0213941 | Sushi domain-containing protein 2 |
| Q6PCB0 | -2.300792 | 0.3926118 | 9.198450 | -5.860221 | 0.0002211 | 0.0249365 | von Willebrand factor A domain-containing protein 1 |
| P35625 | -3.979204 | 0.6344156 | 7.962134 | -6.272235 | 0.0002447 | 0.0261405 | Metalloproteinase inhibitor 3 |
| P60468 | -2.454190 | 0.4052019 | 8.241407 | -6.056708 | 0.0002697 | 0.0268819 | Protein transport protein Sec61 subunit beta |
| Q15327 | -2.221783 | 0.3961366 | 9.280457 | -5.608627 | 0.0002957 | 0.0268819 | Ankyrin repeat domain-containing protein 1 |
| Q14764 | -1.616256 | 0.2901417 | 9.385413 | -5.570573 | 0.0002985 | 0.0268819 | Major vault protein |
| Q69YU5 | 3.220843 | 0.5625486 | 8.841525 | 5.725448 | 0.0003046 | 0.0268819 | Uncharacterized protein C12orf73 |
| Q9P2B2 | -2.045271 | 0.3731311 | 9.385413 | -5.481374 | 0.0003362 | 0.0281811 | Prostaglandin F2 receptor negative regulator |
| P48163 | -2.953929 | 0.4964480 | 7.960353 | -5.950127 | 0.0003486 | 0.0281811 | NADP-dependent malic enzyme |
| Q92930 | -2.803127 | 0.4638105 | 7.597170 | -6.043690 | 0.0003777 | 0.0281811 | Ras-related protein Rab-8B |
| P48681 | -1.493328 | 0.2771541 | 9.321612 | -5.388078 | 0.0003901 | 0.0281811 | Nestin |
| P01031 | -1.652659 | 0.2888520 | 8.183937 | -5.721472 | 0.0004077 | 0.0281811 | Complement C5;Complement C5 beta chain;Complement C5 alpha chain;C5a anaphylatoxin;Complement C5 alpha chain |
| O43677 | -2.965485 | 0.5101963 | 7.933870 | -5.812438 | 0.0004121 | 0.0281811 | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial |
| P02775 | -2.169029 | 0.4075050 | 9.385413 | -5.322705 | 0.0004165 | 0.0281811 | Platelet basic protein;Connective tissue-activating peptide III;TC-2;Connective tissue-activating peptide III(1-81);Beta-thromboglobulin;Neutrophil-activating peptide 2(74);Neutrophil-activating peptide 2(73);Neutrophil-activating peptide 2;TC-1;Neutrophil-activating peptide 2(1-66);Neutrophil-activating peptide 2(1-63) |
| P30711 | -2.323525 | 0.4343764 | 9.018645 | -5.349105 | 0.0004597 | 0.0301015 | Glutathione S-transferase theta-1 |
| Q96FN9 | -2.647962 | 0.4831404 | 8.385413 | -5.480729 | 0.0004988 | 0.0308425 | Probable D-tyrosyl-tRNA(Tyr) deacylase 2 |
| P11586 | 2.011996 | 0.3618415 | 7.881516 | 5.560434 | 0.0005632 | 0.0308425 | C-1-tetrahydrofolate synthase, cytoplasmic;Methylenetetrahydrofolate dehydrogenase;Methenyltetrahydrofolate cyclohydrolase;Formyltetrahydrofolate synthetase;C-1-tetrahydrofolate synthase, cytoplasmic, N-terminally processed |
| Q9H1K0 | -2.283648 | 0.3962045 | 7.385413 | -5.763812 | 0.0005668 | 0.0308425 | Rabenosyn-5 |
| P30405 | -1.828202 | 0.3587630 | 9.385413 | -5.095848 | 0.0005693 | 0.0308425 | Peptidyl-prolyl cis-trans isomerase F, mitochondrial |
| Q09161 | 2.423061 | 0.3818259 | 6.358037 | 6.345983 | 0.0005705 | 0.0308425 | Nuclear cap-binding protein subunit 1 |
| Q5NDL2 | -2.704579 | 0.5131644 | 8.686198 | -5.270394 | 0.0005771 | 0.0308425 | EGF domain-specific O-linked N-acetylglucosamine transferase |
| Q9BZH6 | 3.420486 | 0.6371741 | 8.367230 | 5.368213 | 0.0005773 | 0.0308425 | WD repeat-containing protein 11 |
| O00180 | -4.755346 | 0.8920012 | 8.385413 | -5.331098 | 0.0006003 | 0.0309185 | Potassium channel subfamily K member 1 |
| P00748 | -1.948996 | 0.3807404 | 9.091376 | -5.118963 | 0.0006092 | 0.0309185 | Coagulation factor XII;Coagulation factor XIIa heavy chain;Beta-factor XIIa part 1;Coagulation factor XIIa light chain |
| Q00G26 | 1.936681 | 0.3542707 | 7.788675 | 5.466670 | 0.0006541 | 0.0319953 | Perilipin-5 |
| P23142 | -2.819044 | 0.5094029 | 7.590129 | -5.534018 | 0.0006620 | 0.0319953 | Fibulin-1 |
| P10916 | 2.903591 | 0.5190748 | 7.385413 | 5.593783 | 0.0006814 | 0.0321678 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| Q9UHG2 | -3.305283 | 0.6492888 | 8.796297 | -5.090620 | 0.0007015 | 0.0323665 | ProSAAS;KEP;Big SAAS;Little SAAS;Big PEN-LEN;PEN;Little LEN;Big LEN |
| Q9BW30 | -2.326316 | 0.4679439 | 9.049210 | -4.971357 | 0.0007561 | 0.0336366 | Tubulin polymerization-promoting protein family member 3 |
| P18428 | -1.701410 | 0.3439009 | 9.013977 | -4.947385 | 0.0007905 | 0.0336366 | Lipopolysaccharide-binding protein |
| Q5JPH6 | 3.243753 | 0.5340074 | 6.212234 | 6.074360 | 0.0007941 | 0.0336366 | Probable glutamate–tRNA ligase, mitochondrial |
| Q9BUH6 | -1.516414 | 0.3086486 | 9.115985 | -4.913078 | 0.0008018 | 0.0336366 | Protein PAXX |
| P12883 | 1.752389 | 0.3596357 | 9.255194 | 4.872678 | 0.0008119 | 0.0336366 | Myosin-7 |
| P35052 | -1.955749 | 0.3694381 | 7.703314 | -5.293847 | 0.0008301 | 0.0337017 | Glypican-1;Secreted glypican-1 |
| Q8N474 | -2.161723 | 0.4158032 | 7.847514 | -5.198911 | 0.0008754 | 0.0348447 | Secreted frizzled-related protein 1 |
| Q99983 | -3.574620 | 0.6431377 | 6.904704 | -5.558094 | 0.0008937 | 0.0348873 | Osteomodulin |
| P04004 | -1.932935 | 0.4008337 | 8.981078 | -4.822288 | 0.0009496 | 0.0354254 | Vitronectin;Vitronectin V65 subunit;Vitronectin V10 subunit;Somatomedin-B |
| P62857 | -2.138437 | 0.4200464 | 7.956911 | -5.090954 | 0.0009561 | 0.0354254 | 40S ribosomal protein S28 |
| O15061 | -1.702821 | 0.3601514 | 9.385413 | -4.728071 | 0.0009598 | 0.0354254 | Synemin |
| P08603 | -1.493256 | 0.3168082 | 9.385413 | -4.713438 | 0.0009804 | 0.0355379 | Complement factor H |
| Q9NRG4 | 2.553079 | 0.5223003 | 8.385413 | 4.888143 | 0.0010578 | 0.0376713 | N-lysine methyltransferase SMYD2 |
| Q96KC8 | -2.528784 | 0.5175266 | 8.321700 | -4.886287 | 0.0010840 | 0.0379395 | DnaJ homolog subfamily C member 1 |
| P23434 | 1.413938 | 0.3062675 | 9.385413 | 4.616676 | 0.0011287 | 0.0383884 | Glycine cleavage system H protein, mitochondrial |
| Q04760 | 2.216188 | 0.4645453 | 8.599181 | 4.770661 | 0.0011516 | 0.0383884 | Lactoylglutathione lyase |
| P02452 | -2.007037 | 0.4158054 | 8.367424 | -4.826865 | 0.0011535 | 0.0383884 | Collagen alpha-1(I) chain |
| P24298 | 1.963174 | 0.4165267 | 8.623072 | 4.713201 | 0.0012356 | 0.0398590 | Alanine aminotransferase 1 |
| Q15113 | -1.988509 | 0.4330474 | 9.188121 | -4.591895 | 0.0012370 | 0.0398590 | Procollagen C-endopeptidase enhancer 1 |
| P50453 | -1.422449 | 0.3086989 | 9.052869 | -4.607886 | 0.0012567 | 0.0398605 | Serpin B9 |
| P04275 | -1.362325 | 0.2963324 | 8.991784 | -4.597287 | 0.0012986 | 0.0405566 | von Willebrand factor;von Willebrand antigen 2 |
| Q7L4S7 | -2.547761 | 0.5017188 | 7.157555 | -5.078066 | 0.0013409 | 0.0412425 | Protein ARMCX6 |
| Q86WV6 | -1.336643 | 0.2979661 | 9.239827 | -4.485891 | 0.0014229 | 0.0424928 | Stimulator of interferon genes protein |
| Q92604 | -1.925795 | 0.4331202 | 9.345516 | -4.446330 | 0.0014664 | 0.0424928 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 |
| Q9HCB6 | -2.416374 | 0.5273097 | 8.648303 | -4.582457 | 0.0014674 | 0.0424928 | Spondin-1 |
| P82663 | -1.935089 | 0.4352236 | 9.302714 | -4.446194 | 0.0014834 | 0.0424928 | 28S ribosomal protein S25, mitochondrial |
| Q6PI78 | 1.948479 | 0.4338791 | 9.040261 | 4.490835 | 0.0014919 | 0.0424928 | Transmembrane protein 65 |
| Q8TDB4 | -2.860288 | 0.5377313 | 6.330306 | -5.319177 | 0.0015176 | 0.0424928 | Protein MGARP |
| P01611 | -2.219411 | 0.5030754 | 9.385413 | -4.411687 | 0.0015281 | 0.0424928 | Ig kappa chain V-I region Wes |
| Q15274 | -2.178586 | 0.4620144 | 7.757883 | -4.715407 | 0.0016427 | 0.0426894 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] |
| P08294 | -1.898191 | 0.4077813 | 7.979491 | -4.654924 | 0.0016454 | 0.0426894 | Extracellular superoxide dismutase [Cu-Zn] |
| Q9BSD7 | 2.950896 | 0.6028017 | 7.080846 | 4.895302 | 0.0017057 | 0.0426894 | Cancer-related nucleoside-triphosphatase |
| Q9Y6X5 | -1.544960 | 0.3471001 | 8.750753 | -4.451051 | 0.0017123 | 0.0426894 | Bis(5-adenosyl)-triphosphatase ENPP4 |
| Q6YN16 | 1.546774 | 0.3546814 | 9.222955 | 4.361023 | 0.0017180 | 0.0426894 | Hydroxysteroid dehydrogenase-like protein 2 |
| P01303 | -2.163300 | 0.4894155 | 8.890312 | -4.420171 | 0.0017214 | 0.0426894 | Pro-neuropeptide Y;Neuropeptide Y;C-flanking peptide of NPY |
| P51888 | -1.946837 | 0.3954667 | 6.959924 | -4.922884 | 0.0017365 | 0.0426894 | Prolargin |
| P07360 | -1.547962 | 0.3489250 | 8.752393 | -4.436374 | 0.0017475 | 0.0426894 | Complement component C8 gamma chain |
| P04350 | -2.838038 | 0.5777244 | 6.967833 | -4.912443 | 0.0017514 | 0.0426894 | Tubulin beta-4A chain |
| P24844 | -1.783615 | 0.4121116 | 9.330669 | -4.327990 | 0.0017561 | 0.0426894 | Myosin regulatory light polypeptide 9 |
| Q9UL26 | 7.923835 | 1.7917216 | 8.784585 | 4.422470 | 0.0017665 | 0.0426894 | Ras-related protein Rab-22A |
| P07357 | -1.256415 | 0.2926092 | 9.290174 | -4.293832 | 0.0018671 | 0.0445443 | Complement component C8 alpha chain |
| Q8TBP6 | -1.527281 | 0.3500100 | 8.851865 | -4.363535 | 0.0018871 | 0.0445443 | Solute carrier family 25 member 40 |
| P04003 | -1.392094 | 0.3246379 | 9.090898 | -4.288144 | 0.0019793 | 0.0459247 | C4b-binding protein alpha chain |
| Q6UWS5 | 1.776260 | 0.3726694 | 7.068224 | 4.766316 | 0.0019908 | 0.0459247 | Protein PET117 homolog, mitochondrial |
| O15230 | -1.351267 | 0.3157713 | 9.044210 | -4.279259 | 0.0020293 | 0.0462856 | Laminin subunit alpha-5 |
| O14980 | -1.048446 | 0.2497751 | 9.382448 | -4.197561 | 0.0021118 | 0.0476331 | Exportin-1 |
| O94875-10 | 1.381826 | 0.3251982 | 8.958355 | 4.249182 | 0.0021677 | 0.0483561 | Sorbin and SH3 domain-containing protein 2 |
| P12814 | -2.192490 | 0.5008954 | 8.144626 | -4.377140 | 0.0022587 | 0.0493912 | Alpha-actinin-1 |
| P02743 | -1.660711 | 0.3999916 | 9.385413 | -4.151865 | 0.0022628 | 0.0493912 | Serum amyloid P-component;Serum amyloid P-component(1-203) |
| Q9HAT2 | 1.866882 | 0.4480930 | 9.216677 | 4.166282 | 0.0023025 | 0.0497243 | Sialate O-acetylesterase |
4.5 Evaluate results average contrast \(\log_2 FC_{V-A}\)
4.5.1 Volcano-plot
volcanoAvg <- ggplot(rowData(pe[["proteinRobust"]])$"tissueV + 0.5 * locationR:tissueV",
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) + theme_minimal()
volcanoAvg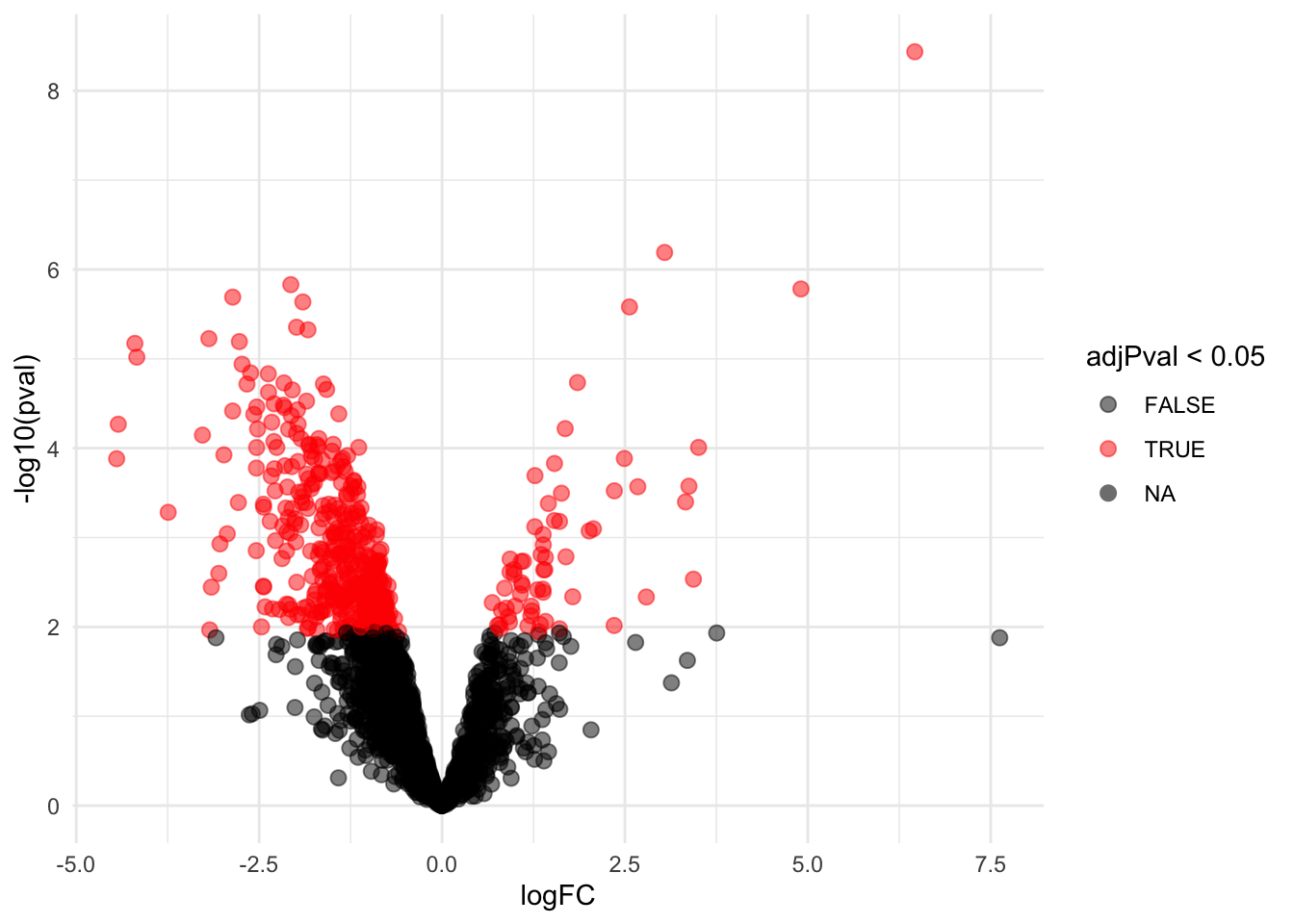
4.5.2 Heatmap
We first select the names of the proteins that were declared signficant.
sigNamesAvg <- rowData(pe[["proteinRobust"]])$"tissueV + 0.5 * locationR:tissueV" %>%
rownames_to_column("proteinRobust") %>%
filter(adjPval<0.05) %>%
pull(proteinRobust)
heatmap(assay(pe[["proteinRobust"]])[sigNamesAvg, ])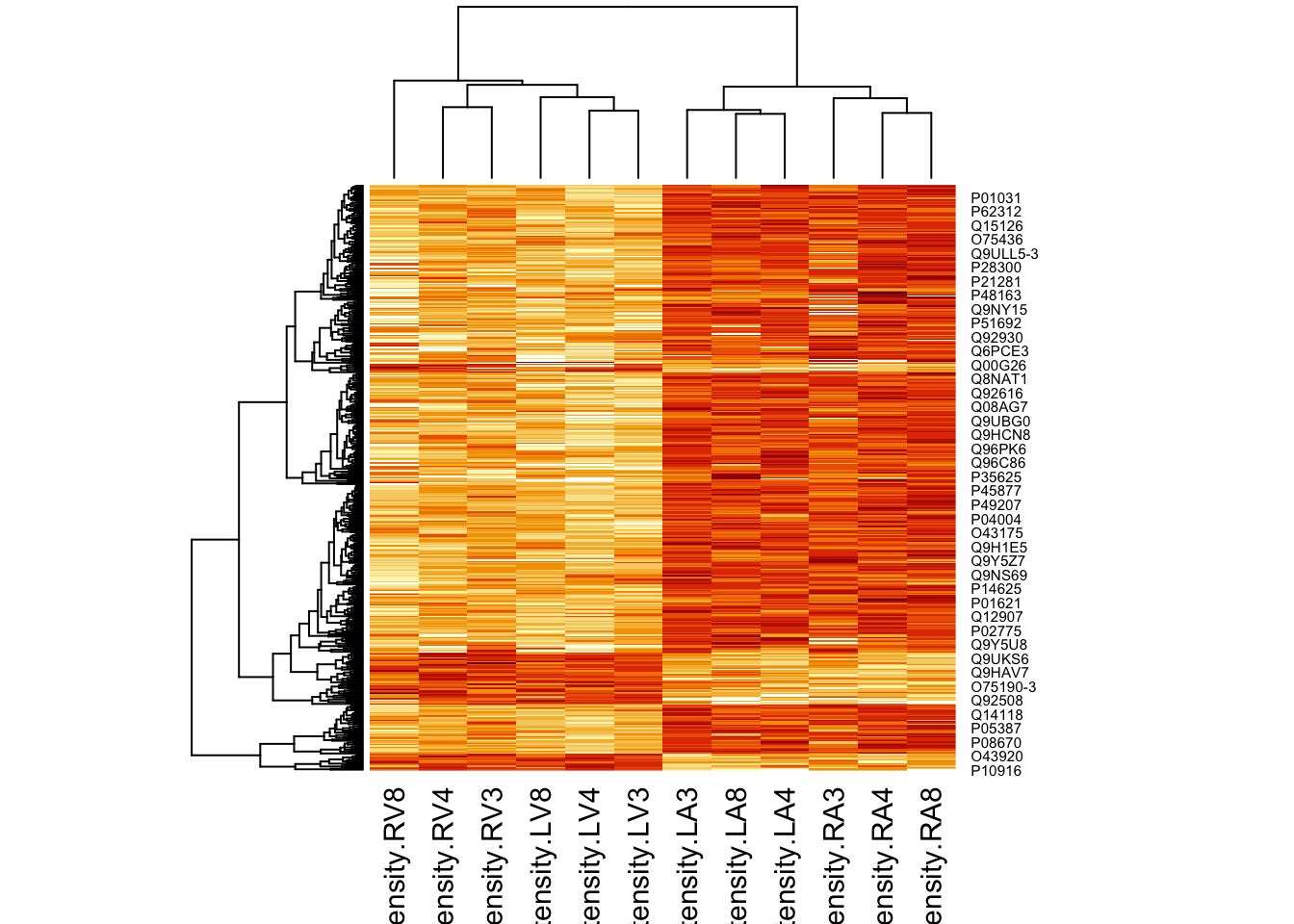
There are 461 proteins significantly differentially expressed at the 5% FDR level.
rowData(pe[["proteinRobust"]])$"tissueV + 0.5 * locationR:tissueV" %>%
cbind(.,rowData(pe[["proteinRobust"]])$Protein.names) %>%
na.exclude %>%
filter(adjPval<0.05) %>%
arrange(pval) %>%
knitr::kable(.)| logFC | se | df | t | pval | adjPval | rowData(pe[[“proteinRobust”]])$Protein.names | |
|---|---|---|---|---|---|---|---|
| P08590 | 6.4625112 | 0.2976280 | 9.117912 | 21.713386 | 0.0000000 | 0.0000074 | Myosin light chain 3 |
| P12883 | 3.0427569 | 0.2557724 | 9.255194 | 11.896344 | 0.0000006 | 0.0006551 | Myosin-7 |
| O75368 | -2.0672256 | 0.1911057 | 9.251939 | -10.817185 | 0.0000015 | 0.0007617 | SH3 domain-binding glutamic acid-rich-like protein |
| P10916 | 4.9061329 | 0.3591470 | 7.385413 | 13.660515 | 0.0000017 | 0.0007617 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| Q6UWY5 | -2.8614477 | 0.2723232 | 9.170135 | -10.507543 | 0.0000020 | 0.0007617 | Olfactomedin-like protein 1 |
| P46821 | -1.9028923 | 0.1876947 | 9.385413 | -10.138230 | 0.0000023 | 0.0007617 | Microtubule-associated protein 1B;MAP1B heavy chain;MAP1 light chain LC1 |
| P06858 | 2.5622451 | 0.2524054 | 9.218266 | 10.151307 | 0.0000026 | 0.0007617 | Lipoprotein lipase |
| P29622 | -1.9885126 | 0.2115311 | 9.385413 | -9.400570 | 0.0000044 | 0.0010675 | Kallistatin |
| P05546 | -1.8329275 | 0.1931907 | 9.198639 | -9.487658 | 0.0000047 | 0.0010675 | Heparin cofactor 2 |
| P21810 | -3.1872304 | 0.3151219 | 8.304055 | -10.114278 | 0.0000059 | 0.0011366 | Biglycan |
| P35442 | -2.7706752 | 0.2792024 | 8.385413 | -9.923538 | 0.0000064 | 0.0011366 | Thrombospondin-2 |
| P13533 | -4.1989377 | 0.4579673 | 9.115028 | -9.168640 | 0.0000067 | 0.0011366 | Myosin-6 |
| Q06828 | -4.1723327 | 0.4703068 | 9.003303 | -8.871513 | 0.0000096 | 0.0014958 | Fibromodulin |
| Q8N474 | -2.7331853 | 0.2795787 | 7.847514 | -9.776084 | 0.0000115 | 0.0016658 | Secreted frizzled-related protein 1 |
| P02452 | -2.6166150 | 0.2921724 | 8.367424 | -8.955722 | 0.0000144 | 0.0018663 | Collagen alpha-1(I) chain |
| Q9UGT4 | -2.3750749 | 0.2867846 | 9.193722 | -8.281739 | 0.0000147 | 0.0018663 | Sushi domain-containing protein 2 |
| O94875-10 | 1.8515322 | 0.2254187 | 8.958355 | 8.213746 | 0.0000184 | 0.0019441 | Sorbin and SH3 domain-containing protein 2 |
| Q16647 | -2.1609826 | 0.2677306 | 9.156725 | -8.071483 | 0.0000186 | 0.0019441 | Prostacyclin synthase |
| Q14764 | -1.6211158 | 0.2051612 | 9.385413 | -7.901669 | 0.0000191 | 0.0019441 | Major vault protein |
| A6NMZ7 | -2.6666436 | 0.3375438 | 9.385413 | -7.900140 | 0.0000192 | 0.0019441 | Collagen alpha-6(VI) chain |
| O95865 | -1.5774343 | 0.2030636 | 9.385413 | -7.768178 | 0.0000220 | 0.0020570 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 |
| Q9P2B2 | -2.0463902 | 0.2638435 | 9.385413 | -7.756076 | 0.0000223 | 0.0020570 | Prostaglandin F2 receptor negative regulator |
| Q15113 | -2.3717647 | 0.3035044 | 9.188121 | -7.814597 | 0.0000237 | 0.0020912 | Procollagen C-endopeptidase enhancer 1 |
| P18428 | -1.8519938 | 0.2405365 | 9.013977 | -7.699428 | 0.0000298 | 0.0025169 | Lipopolysaccharide-binding protein |
| P02776 | -2.2952145 | 0.2665516 | 7.718405 | -8.610771 | 0.0000319 | 0.0025481 | Platelet factor 4;Platelet factor 4, short form |
| P24844 | -2.1677081 | 0.2920895 | 9.330669 | -7.421383 | 0.0000330 | 0.0025481 | Myosin regulatory light polypeptide 9 |
| Q9BW30 | -2.5325002 | 0.3361911 | 9.049210 | -7.532919 | 0.0000346 | 0.0025481 | Tubulin polymerization-promoting protein family member 3 |
| P54652 | -2.1555836 | 0.2857710 | 9.007166 | -7.543045 | 0.0000351 | 0.0025481 | Heat shock-related 70 kDa protein 2 |
| P00748 | -1.9760261 | 0.2655421 | 9.091376 | -7.441479 | 0.0000372 | 0.0025980 | Coagulation factor XII;Coagulation factor XIIa heavy chain;Beta-factor XIIa part 1;Coagulation factor XIIa light chain |
| O43677 | -2.8617874 | 0.3490622 | 7.933870 | -8.198503 | 0.0000384 | 0.0025980 | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial |
| P48681 | -1.4117043 | 0.1954445 | 9.321612 | -7.223044 | 0.0000413 | 0.0026554 | Nestin |
| P51888 | -2.5731928 | 0.2835498 | 6.959924 | -9.074923 | 0.0000419 | 0.0026554 | Prolargin |
| P02775 | -2.0605776 | 0.2881496 | 9.385413 | -7.151070 | 0.0000432 | 0.0026568 | Platelet basic protein;Connective tissue-activating peptide III;TC-2;Connective tissue-activating peptide III(1-81);Beta-thromboglobulin;Neutrophil-activating peptide 2(74);Neutrophil-activating peptide 2(73);Neutrophil-activating peptide 2;TC-1;Neutrophil-activating peptide 2(1-66);Neutrophil-activating peptide 2(1-63) |
| P08294 | -2.3273246 | 0.2968595 | 7.979491 | -7.839819 | 0.0000512 | 0.0030425 | Extracellular superoxide dismutase [Cu-Zn] |
| O00180 | -4.4260339 | 0.5900034 | 8.385413 | -7.501710 | 0.0000539 | 0.0030425 | Potassium channel subfamily K member 1 |
| P02743 | -1.9672445 | 0.2828368 | 9.385413 | -6.955406 | 0.0000540 | 0.0030425 | Serum amyloid P-component;Serum amyloid P-component(1-203) |
| P14854 | 1.6832743 | 0.2271074 | 8.346841 | 7.411798 | 0.0000604 | 0.0031941 | Cytochrome c oxidase subunit 6B1 |
| Q5NDL2 | -2.5211042 | 0.3501742 | 8.686198 | -7.199572 | 0.0000610 | 0.0031941 | EGF domain-specific O-linked N-acetylglucosamine transferase |
| Q92604 | -2.0980384 | 0.3057458 | 9.345516 | -6.862036 | 0.0000614 | 0.0031941 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 |
| P60468 | -1.9885569 | 0.2703727 | 8.241407 | -7.354874 | 0.0000683 | 0.0034653 | Protein transport protein Sec61 subunit beta |
| P23083 | -3.2749341 | 0.4122019 | 7.385413 | -7.944976 | 0.0000714 | 0.0035334 | Ig heavy chain V-I region V35 |
| P30405 | -1.6842375 | 0.2536837 | 9.385413 | -6.639124 | 0.0000781 | 0.0037005 | Peptidyl-prolyl cis-trans isomerase F, mitochondrial |
| P51884 | -1.9277723 | 0.2600484 | 7.936878 | -7.413128 | 0.0000784 | 0.0037005 | Lumican |
| Q9ULL5-3 | -2.2984552 | 0.3119661 | 7.893242 | -7.367645 | 0.0000842 | 0.0037660 | Proline-rich protein 12 |
| P01031 | -1.4875185 | 0.2092918 | 8.183937 | -7.107390 | 0.0000905 | 0.0037660 | Complement C5;Complement C5 beta chain;Complement C5 alpha chain;C5a anaphylatoxin;Complement C5 alpha chain |
| Q6PCB0 | -1.8152862 | 0.2753405 | 9.198450 | -6.592879 | 0.0000905 | 0.0037660 | von Willebrand factor A domain-containing protein 1 |
| Q8TBP6 | -1.7102618 | 0.2536922 | 8.851865 | -6.741483 | 0.0000914 | 0.0037660 | Solute carrier family 25 member 40 |
| Q53GQ0 | -1.8232291 | 0.2797866 | 9.334954 | -6.516498 | 0.0000926 | 0.0037660 | Very-long-chain 3-oxoacyl-CoA reductase |
| P36955 | -1.6946775 | 0.2459657 | 8.456893 | -6.889895 | 0.0000966 | 0.0037660 | Pigment epithelium-derived factor |
| O14980 | -1.1393991 | 0.1766395 | 9.382448 | -6.450420 | 0.0000980 | 0.0037660 | Exportin-1 |
| Q92508 | 3.5066909 | 0.4088131 | 6.385413 | 8.577735 | 0.0000980 | 0.0037660 | Piezo-type mechanosensitive ion channel component 1 |
| Q14195-2 | -2.2588828 | 0.3476626 | 9.262403 | -6.497341 | 0.0000982 | 0.0037660 | Dihydropyrimidinase-related protein 3 |
| P05997 | -2.5339895 | 0.3799038 | 8.864434 | -6.670083 | 0.0000983 | 0.0037660 | Collagen alpha-2(V) chain |
| Q15327 | -1.7881570 | 0.2788386 | 9.280457 | -6.412874 | 0.0001077 | 0.0039884 | Ankyrin repeat domain-containing protein 1 |
| P00325 | -1.5039787 | 0.2325735 | 9.141096 | -6.466682 | 0.0001081 | 0.0039884 | Alcohol dehydrogenase 1B |
| P35625 | -2.9820031 | 0.4278849 | 7.962134 | -6.969171 | 0.0001188 | 0.0043077 | Metalloproteinase inhibitor 3 |
| P07451 | -1.2910732 | 0.2057778 | 9.385413 | -6.274113 | 0.0001214 | 0.0043228 | Carbonic anhydrase 3 |
| P04004 | -1.7834542 | 0.2780483 | 8.981078 | -6.414189 | 0.0001244 | 0.0043267 | Vitronectin;Vitronectin V65 subunit;Vitronectin V10 subunit;Somatomedin-B |
| P41240 | -1.3613815 | 0.2127198 | 8.922337 | -6.399880 | 0.0001302 | 0.0043267 | Tyrosine-protein kinase CSK |
| Q69YU5 | 2.4923143 | 0.3874663 | 8.841525 | 6.432339 | 0.0001305 | 0.0043267 | Uncharacterized protein C12orf73 |
| Q8WWA0 | -4.4480904 | 0.6626292 | 8.244689 | -6.712790 | 0.0001313 | 0.0043267 | Intelectin-1 |
| P46060 | -1.7409587 | 0.2624323 | 8.385413 | -6.633934 | 0.0001321 | 0.0043267 | Ran GTPase-activating protein 1 |
| P28066 | -1.6401936 | 0.2502063 | 8.500773 | -6.555364 | 0.0001353 | 0.0043611 | Proteasome subunit alpha type-5 |
| O15230 | -1.3831391 | 0.2198154 | 9.044210 | -6.292275 | 0.0001393 | 0.0044015 | Laminin subunit alpha-5 |
| O95980 | -1.9680314 | 0.2898803 | 7.980660 | -6.789117 | 0.0001409 | 0.0044015 | Reversion-inducing cysteine-rich protein with Kazal motifs |
| Q6YN16 | 1.5369703 | 0.2490340 | 9.222955 | 6.171729 | 0.0001483 | 0.0045619 | Hydroxysteroid dehydrogenase-like protein 2 |
| Q7L4S7 | -2.1476054 | 0.2976126 | 7.157555 | -7.216110 | 0.0001572 | 0.0046901 | Protein ARMCX6 |
| P04003 | -1.4355734 | 0.2328336 | 9.090898 | -6.165662 | 0.0001587 | 0.0046901 | C4b-binding protein alpha chain |
| Q15274 | -2.0534369 | 0.3025225 | 7.757883 | -6.787717 | 0.0001608 | 0.0046901 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] |
| P04083 | -1.3497192 | 0.2233143 | 9.385413 | -6.044035 | 0.0001617 | 0.0046901 | Annexin A1 |
| P36021 | -2.5372663 | 0.3952239 | 8.385413 | -6.419821 | 0.0001670 | 0.0047747 | Monocarboxylate transporter 8 |
| Q9BXN1 | -2.2900275 | 0.3712225 | 8.937487 | -6.168882 | 0.0001698 | 0.0047874 | Asporin |
| Q9Y6X5 | -1.4781975 | 0.2387999 | 8.750753 | -6.190111 | 0.0001809 | 0.0050078 | Bis(5-adenosyl)-triphosphatase ENPP4 |
| Q07954 | -1.3233833 | 0.2224711 | 9.385413 | -5.948564 | 0.0001826 | 0.0050078 | Prolow-density lipoprotein receptor-related protein 1;Low-density lipoprotein receptor-related protein 1 85 kDa subunit;Low-density lipoprotein receptor-related protein 1 515 kDa subunit;Low-density lipoprotein receptor-related protein 1 intracellular domain |
| O75828 | -1.5053475 | 0.2540246 | 9.385413 | -5.925992 | 0.0001879 | 0.0050083 | Carbonyl reductase [NADPH] 3 |
| P12110 | -1.6865779 | 0.2700881 | 8.535409 | -6.244548 | 0.0001885 | 0.0050083 | Collagen alpha-2(VI) chain |
| Q9UBG0 | -1.6619433 | 0.2773921 | 9.131671 | -5.991314 | 0.0001931 | 0.0050083 | C-type mannose receptor 2 |
| P04196 | -1.6884017 | 0.2542452 | 7.683140 | -6.640839 | 0.0001946 | 0.0050083 | Histidine-rich glycoprotein |
| Q9NZ01 | -1.8500678 | 0.3007922 | 8.691035 | -6.150652 | 0.0001949 | 0.0050083 | Very-long-chain enoyl-CoA reductase |
| P23434 | 1.2703798 | 0.2165638 | 9.385413 | 5.866076 | 0.0002029 | 0.0051267 | Glycine cleavage system H protein, mitochondrial |
| P23142 | -2.3359021 | 0.3515315 | 7.590129 | -6.644929 | 0.0002046 | 0.0051267 | Fibulin-1 |
| Q92681 | -1.8263794 | 0.2904491 | 8.133279 | -6.288122 | 0.0002200 | 0.0054460 | Regulatory solute carrier protein family 1 member 1 |
| P07357 | -1.2056635 | 0.2077663 | 9.290174 | -5.802980 | 0.0002289 | 0.0055985 | Complement component C8 alpha chain |
| P04275 | -1.2082122 | 0.2055449 | 8.991784 | -5.878095 | 0.0002362 | 0.0057087 | von Willebrand factor;von Willebrand antigen 2 |
| P49207 | -1.3752484 | 0.2347644 | 8.970773 | -5.857995 | 0.0002444 | 0.0058016 | 60S ribosomal protein L34 |
| Q8TBQ9 | -1.7430434 | 0.2824761 | 8.170120 | -6.170588 | 0.0002458 | 0.0058016 | Protein kish-A |
| Q9BUF5 | -1.7774391 | 0.3096233 | 9.217423 | -5.740650 | 0.0002555 | 0.0059622 | Tubulin beta-6 chain |
| P50453 | -1.2397969 | 0.2150088 | 9.052869 | -5.766260 | 0.0002648 | 0.0060077 | Serpin B9 |
| Q5JPH6 | 3.3748852 | 0.4562924 | 6.212234 | 7.396321 | 0.0002665 | 0.0060077 | Probable glutamate–tRNA ligase, mitochondrial |
| Q9BZH6 | 2.6758207 | 0.4457948 | 8.367230 | 6.002360 | 0.0002702 | 0.0060077 | WD repeat-containing protein 11 |
| P01024 | -1.1496569 | 0.2015704 | 9.197920 | -5.703501 | 0.0002702 | 0.0060077 | Complement C3;Complement C3 beta chain;C3-beta-c;Complement C3 alpha chain;C3a anaphylatoxin;Acylation stimulating protein;Complement C3b alpha chain;Complement C3c alpha chain fragment 1;Complement C3dg fragment;Complement C3g fragment;Complement C3d fragment;Complement C3f fragment;Complement C3c alpha chain fragment 2 |
| Q9HCB6 | -2.1126269 | 0.3589347 | 8.648303 | -5.885825 | 0.0002723 | 0.0060077 | Spondin-1 |
| Q86WV6 | -1.2006082 | 0.2120678 | 9.239827 | -5.661436 | 0.0002805 | 0.0061226 | Stimulator of interferon genes protein |
| P30711 | -1.7885395 | 0.3126012 | 9.018645 | -5.721474 | 0.0002842 | 0.0061378 | Glutathione S-transferase theta-1 |
| Q9NRG4 | 2.3553762 | 0.3989134 | 8.385413 | 5.904480 | 0.0003002 | 0.0063497 | N-lysine methyltransferase SMYD2 |
| O60760 | -2.2815940 | 0.3864378 | 8.385413 | -5.904169 | 0.0003003 | 0.0063497 | Hematopoietic prostaglandin D synthase |
| P62760 | -1.8910941 | 0.3355983 | 9.120970 | -5.634992 | 0.0003044 | 0.0063707 | Visinin-like protein 1 |
| P12814 | -1.9593420 | 0.3281249 | 8.144626 | -5.971329 | 0.0003113 | 0.0064488 | Alpha-actinin-1 |
| P24298 | 1.6321192 | 0.2831640 | 8.623072 | 5.763865 | 0.0003191 | 0.0065425 | Alanine aminotransferase 1 |
| Q9UQ35 | -1.2479485 | 0.2265532 | 9.385413 | -5.508412 | 0.0003242 | 0.0065816 | Serine/arginine repetitive matrix protein 2 |
| P14550 | -1.3044364 | 0.2225655 | 8.301450 | -5.860910 | 0.0003283 | 0.0065991 | Alcohol dehydrogenase [NADP(+)] |
| O94919 | -1.1546247 | 0.2088661 | 9.248102 | -5.528061 | 0.0003329 | 0.0066246 | Endonuclease domain-containing 1 protein |
| Q9Y3B4 | -1.2989386 | 0.2361784 | 9.218746 | -5.499820 | 0.0003494 | 0.0068655 | Splicing factor 3B subunit 6 |
| P49458 | -1.9200693 | 0.3013686 | 7.120983 | -6.371165 | 0.0003517 | 0.0068655 | Signal recognition particle 9 kDa protein |
| Q9UNW9 | 3.3281626 | 0.6111632 | 9.054127 | 5.445620 | 0.0003996 | 0.0076489 | RNA-binding protein Nova-2 |
| P48163 | -1.9140897 | 0.3290692 | 7.960353 | -5.816679 | 0.0004050 | 0.0076489 | NADP-dependent malic enzyme |
| Q5M9N0 | -2.7847331 | 0.4916908 | 8.350040 | -5.663586 | 0.0004057 | 0.0076489 | Coiled-coil domain-containing protein 158 |
| P02747 | -1.8649499 | 0.3002262 | 7.138784 | -6.211815 | 0.0004069 | 0.0076489 | Complement C1q subcomponent subunit C |
| Q00G26 | 1.4536200 | 0.2477814 | 7.788675 | 5.866543 | 0.0004159 | 0.0077448 | Perilipin-5 |
| Q8WY22 | -1.5477211 | 0.2756010 | 8.385413 | -5.615805 | 0.0004231 | 0.0077538 | BRI3-binding protein |
| Q9UHG2 | -2.4417072 | 0.4457629 | 8.796297 | -5.477592 | 0.0004240 | 0.0077538 | ProSAAS;KEP;Big SAAS;Little SAAS;Big PEN-LEN;PEN;Little LEN;Big LEN |
| Q9ULC3 | -1.4634583 | 0.2660801 | 8.669265 | -5.500068 | 0.0004336 | 0.0078588 | Ras-related protein Rab-23 |
| P25940 | -1.6268129 | 0.3026580 | 9.043783 | -5.375087 | 0.0004401 | 0.0079054 | Collagen alpha-3(V) chain |
| Q53GG5-2 | -2.4390475 | 0.4404803 | 8.429675 | -5.537246 | 0.0004569 | 0.0080899 | PDZ and LIM domain protein 3 |
| Q8WZA9 | -1.1072814 | 0.2082637 | 9.101344 | -5.316729 | 0.0004653 | 0.0080899 | Immunity-related GTPase family Q protein |
| P07585 | -2.1345776 | 0.4031029 | 9.163968 | -5.295366 | 0.0004680 | 0.0080899 | Decorin |
| O00264 | -1.4169853 | 0.2667510 | 9.079390 | -5.312014 | 0.0004721 | 0.0080899 | Membrane-associated progesterone receptor component 1 |
| Q53T59 | -1.8337712 | 0.3208050 | 7.876061 | -5.716155 | 0.0004722 | 0.0080899 | HCLS1-binding protein 3 |
| Q6SZW1 | -2.0501728 | 0.3456301 | 7.385413 | -5.931696 | 0.0004742 | 0.0080899 | Sterile alpha and TIR motif-containing protein 1 |
| Q9BS26 | -1.1652487 | 0.2228454 | 9.249914 | -5.228955 | 0.0004964 | 0.0083980 | Endoplasmic reticulum resident protein 44 |
| P01008 | -1.5396740 | 0.2964219 | 9.310907 | -5.194199 | 0.0005096 | 0.0085495 | Antithrombin-III |
| Q9UKR5 | -3.7435260 | 0.6964057 | 8.595465 | -5.375496 | 0.0005225 | 0.0086032 | Probable ergosterol biosynthetic protein 28 |
| P14543 | -1.3422700 | 0.2580050 | 9.204669 | -5.202496 | 0.0005229 | 0.0086032 | Nidogen-1 |
| P08603 | -1.1544473 | 0.2240173 | 9.385413 | -5.153386 | 0.0005255 | 0.0086032 | Complement factor H |
| Q92621 | -1.2423196 | 0.2415233 | 9.385413 | -5.143684 | 0.0005326 | 0.0086499 | Nuclear pore complex protein Nup205 |
| Q9BTV4 | -1.6057405 | 0.3110895 | 9.284468 | -5.161667 | 0.0005377 | 0.0086637 | Transmembrane protein 43 |
| P13671 | -1.3648753 | 0.2664751 | 9.385413 | -5.121963 | 0.0005489 | 0.0087744 | Complement component C6 |
| Q2TAA5 | -1.1443503 | 0.2235983 | 9.249693 | -5.117885 | 0.0005779 | 0.0091655 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase |
| Q12996 | -1.4804805 | 0.2552304 | 7.207709 | -5.800565 | 0.0005960 | 0.0093205 | Cleavage stimulation factor subunit 3 |
| P20774 | -2.1010556 | 0.3896021 | 8.210882 | -5.392824 | 0.0005969 | 0.0093205 | Mimecan |
| Q9UJC5 | -1.2259781 | 0.2425389 | 9.326832 | -5.054769 | 0.0006146 | 0.0094299 | SH3 domain-binding glutamic acid-rich-like protein 2 |
| P01042 | -2.0066697 | 0.3480072 | 7.210294 | -5.766173 | 0.0006170 | 0.0094299 | Kininogen-1;Kininogen-1 heavy chain;T-kinin;Bradykinin;Lysyl-bradykinin;Kininogen-1 light chain;Low molecular weight growth-promoting factor |
| O14967 | -1.6369952 | 0.3092277 | 8.430438 | -5.293818 | 0.0006179 | 0.0094299 | Calmegin |
| Q16082 | -1.3233427 | 0.2607936 | 9.207589 | -5.074291 | 0.0006225 | 0.0094299 | Heat shock protein beta-2 |
| Q6PI78 | 1.5357334 | 0.3017719 | 9.040261 | 5.089054 | 0.0006457 | 0.0097101 | Transmembrane protein 65 |
| Q8TDB4 | -2.3494772 | 0.3787763 | 6.330306 | -6.202809 | 0.0006590 | 0.0097915 | Protein MGARP |
| Q9HAT2 | 1.6055669 | 0.3192733 | 9.216677 | 5.028817 | 0.0006608 | 0.0097915 | Sialate O-acetylesterase |
| Q9BUH6 | -1.1106212 | 0.2210609 | 9.115985 | -5.024051 | 0.0006878 | 0.0100890 | Protein PAXX |
| Q9ULD0 | -2.0151143 | 0.3202146 | 6.128068 | -6.293011 | 0.0006908 | 0.0100890 | 2-oxoglutarate dehydrogenase-like, mitochondrial |
| Q14314 | -1.9322639 | 0.3920483 | 9.382448 | -4.928637 | 0.0007208 | 0.0104515 | Fibroleukin |
| P45877 | -0.9996304 | 0.2027328 | 9.343571 | -4.930777 | 0.0007275 | 0.0104735 | Peptidyl-prolyl cis-trans isomerase C |
| Q96H79 | -2.1774013 | 0.3587038 | 6.334231 | -6.070192 | 0.0007408 | 0.0105623 | Zinc finger CCCH-type antiviral protein 1-like |
| P35052 | -1.2875376 | 0.2389775 | 7.703314 | -5.387694 | 0.0007440 | 0.0105623 | Glypican-1;Secreted glypican-1 |
| P11586 | 1.2680380 | 0.2388823 | 7.881516 | 5.308212 | 0.0007574 | 0.0106771 | C-1-tetrahydrofolate synthase, cytoplasmic;Methylenetetrahydrofolate dehydrogenase;Methenyltetrahydrofolate cyclohydrolase;Formyltetrahydrofolate synthetase;C-1-tetrahydrofolate synthase, cytoplasmic, N-terminally processed |
| Q9HAV4 | -1.6824690 | 0.3206960 | 7.987893 | -5.246305 | 0.0007811 | 0.0109357 | Exportin-5 |
| P40261 | -1.4339415 | 0.2942762 | 9.340024 | -4.872774 | 0.0007907 | 0.0109462 | Nicotinamide N-methyltransferase |
| P02790 | -1.3244981 | 0.2724643 | 9.385413 | -4.861179 | 0.0007927 | 0.0109462 | Hemopexin |
| P19429 | 2.0691315 | 0.3954475 | 7.976624 | 5.232379 | 0.0007980 | 0.0109462 | Troponin I, cardiac muscle |
| P34932 | -0.8925962 | 0.1844794 | 9.385413 | -4.838461 | 0.0008188 | 0.0111559 | Heat shock 70 kDa protein 4 |
| Q09161 | 2.0115745 | 0.3402010 | 6.358037 | 5.912900 | 0.0008442 | 0.0114245 | Nuclear cap-binding protein subunit 1 |
| Q15126 | -1.0103044 | 0.2097310 | 9.342811 | -4.817144 | 0.0008552 | 0.0114968 | Phosphomevalonate kinase |
| P50479 | -2.1200408 | 0.4140571 | 8.131897 | -5.120165 | 0.0008621 | 0.0115130 | PDZ and LIM domain protein 4 |
| Q9NY15 | -1.4835786 | 0.3089047 | 9.336638 | -4.802707 | 0.0008747 | 0.0116049 | Stabilin-1 |
| Q86VP6 | -1.3548284 | 0.2717978 | 8.542953 | -4.984693 | 0.0008822 | 0.0116274 | Cullin-associated NEDD8-dissociated protein 1 |
| Q07507 | -1.3153838 | 0.2587790 | 8.160106 | -5.083039 | 0.0008935 | 0.0116274 | Dermatopontin |
| O43175 | -2.0775407 | 0.4307821 | 9.173117 | -4.822719 | 0.0008937 | 0.0116274 | D-3-phosphoglycerate dehydrogenase |
| P02461 | -2.9357775 | 0.5844720 | 8.323445 | -5.022957 | 0.0009077 | 0.0116274 | Collagen alpha-1(III) chain |
| P01034 | -1.4660858 | 0.3001221 | 8.837162 | -4.884964 | 0.0009125 | 0.0116274 | Cystatin-C |
| P28300 | -1.4793279 | 0.2827310 | 7.649016 | -5.232280 | 0.0009129 | 0.0116274 | Protein-lysine 6-oxidase |
| P13667 | -1.1046003 | 0.2299757 | 9.181460 | -4.803117 | 0.0009164 | 0.0116274 | Protein disulfide-isomerase A4 |
| Q86VU5 | 1.3820110 | 0.2907797 | 9.385413 | 4.752776 | 0.0009261 | 0.0116774 | Catechol O-methyltransferase domain-containing protein 1 |
| O43143 | -0.8972350 | 0.1881734 | 9.289616 | -4.768130 | 0.0009321 | 0.0116801 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 |
| O95183 | -1.4595318 | 0.3089309 | 9.371170 | -4.724460 | 0.0009689 | 0.0120661 | Vesicle-associated membrane protein 5 |
| Q96CS3 | -1.1904104 | 0.2496881 | 9.119924 | -4.767590 | 0.0009819 | 0.0121212 | FAS-associated factor 2 |
| P06727 | -1.0625381 | 0.2206360 | 8.884167 | -4.815797 | 0.0009881 | 0.0121212 | Apolipoprotein A-IV |
| P09619 | -1.2925211 | 0.2721302 | 9.154490 | -4.749643 | 0.0009967 | 0.0121212 | Platelet-derived growth factor receptor beta |
| P26447 | -1.5232285 | 0.3150582 | 8.774968 | -4.834753 | 0.0009972 | 0.0121212 | Protein S100-A4 |
| P46940 | -1.1176083 | 0.2393244 | 9.306592 | -4.669847 | 0.0010683 | 0.0129091 | Ras GTPase-activating-like protein IQGAP1 |
| Q15582 | -2.2755713 | 0.4543685 | 7.899813 | -5.008206 | 0.0010822 | 0.0129993 | Transforming growth factor-beta-induced protein ig-h3 |
| Q96JB2 | -1.9983947 | 0.4070184 | 8.142504 | -4.909838 | 0.0011200 | 0.0133746 | Conserved oligomeric Golgi complex subunit 3 |
| P01019 | -1.0245069 | 0.2195559 | 9.093510 | -4.666269 | 0.0011427 | 0.0135650 | Angiotensinogen;Angiotensin-1;Angiotensin-2;Angiotensin-3;Angiotensin-4;Angiotensin 1-9;Angiotensin 1-7;Angiotensin 1-5;Angiotensin 1-4 |
| Q99983 | -3.0353776 | 0.5731048 | 6.904704 | -5.296374 | 0.0011784 | 0.0139082 | Osteomodulin |
| Q96LL9 | -1.2761049 | 0.2726611 | 8.842792 | -4.680187 | 0.0012079 | 0.0140956 | DnaJ homolog subfamily C member 30 |
| Q9NRX4 | 1.3837621 | 0.3021988 | 9.339127 | 4.578979 | 0.0012082 | 0.0140956 | 14 kDa phosphohistidine phosphatase |
| O95486 | -1.3108014 | 0.2851357 | 9.187800 | -4.597114 | 0.0012278 | 0.0142428 | Protein transport protein Sec24A |
| P62328 | -1.4789049 | 0.2892444 | 7.220282 | -5.112994 | 0.0012549 | 0.0144745 | Thymosin beta-4;Hematopoietic system regulatory peptide |
| P02748 | -1.2312031 | 0.2668957 | 8.865762 | -4.613050 | 0.0013178 | 0.0151138 | Complement component C9;Complement component C9a;Complement component C9b |
| P83916 | -1.6679489 | 0.3077674 | 6.385413 | -5.419511 | 0.0013357 | 0.0152331 | Chromobox protein homolog 1 |
| Q8WWQ0 | -1.2921726 | 0.2873956 | 9.385413 | -4.496147 | 0.0013478 | 0.0152848 | PH-interacting protein |
| P00747 | -0.8346905 | 0.1843299 | 9.172800 | -4.528243 | 0.0013623 | 0.0153642 | Plasminogen;Plasmin heavy chain A;Activation peptide;Angiostatin;Plasmin heavy chain A, short form;Plasmin light chain B |
| P27658 | -1.6429540 | 0.3664317 | 9.385413 | -4.483656 | 0.0013729 | 0.0153982 | Collagen alpha-1(VIII) chain;Vastatin |
| O95445 | -2.5390830 | 0.5603209 | 9.052359 | -4.531480 | 0.0014023 | 0.0154891 | Apolipoprotein M |
| Q9UBV8 | -1.1721927 | 0.2568550 | 8.885488 | -4.563635 | 0.0014048 | 0.0154891 | Peflin |
| Q92930 | -1.4926298 | 0.3053336 | 7.597170 | -4.888521 | 0.0014055 | 0.0154891 | Ras-related protein Rab-8B |
| O15118 | -2.1251890 | 0.3964131 | 6.385413 | -5.361047 | 0.0014158 | 0.0154891 | Niemann-Pick C1 protein |
| P03950 | -1.7958993 | 0.3814592 | 8.216113 | -4.707972 | 0.0014192 | 0.0154891 | Angiogenin |
| Q8NAT1 | -0.8669596 | 0.1945741 | 9.385413 | -4.455677 | 0.0014312 | 0.0155361 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 |
| Q9Y287 | -1.6263595 | 0.3659544 | 9.385413 | -4.444159 | 0.0014559 | 0.0157204 | Integral membrane protein 2B;BRI2, membrane form;BRI2 intracellular domain;BRI2C, soluble form;Bri23 peptide |
| Q7Z3T8 | -1.3963655 | 0.3020251 | 8.413206 | -4.623343 | 0.0014913 | 0.0159618 | Zinc finger FYVE domain-containing protein 16 |
| P62857 | -1.3918700 | 0.2939084 | 7.956911 | -4.735728 | 0.0014940 | 0.0159618 | 40S ribosomal protein S28 |
| Q08945 | -1.4241196 | 0.3089376 | 8.385413 | -4.609731 | 0.0015322 | 0.0162239 | FACT complex subunit SSRP1 |
| P82663 | -1.3644647 | 0.3087747 | 9.302714 | -4.418965 | 0.0015444 | 0.0162239 | 28S ribosomal protein S25, mitochondrial |
| Q9BYN0 | -1.2858687 | 0.2896091 | 9.180640 | -4.440015 | 0.0015462 | 0.0162239 | Sulfiredoxin-1 |
| Q6UWS5 | 1.3552496 | 0.2719635 | 7.068224 | 4.983204 | 0.0015505 | 0.0162239 | Protein PET117 homolog, mitochondrial |
| P54577 | -1.0275324 | 0.2300731 | 8.964139 | -4.466112 | 0.0015790 | 0.0163449 | Tyrosine–tRNA ligase, cytoplasmic;Tyrosine–tRNA ligase, cytoplasmic, N-terminally processed |
| P00352 | -1.1336794 | 0.2474061 | 8.402816 | -4.582262 | 0.0015817 | 0.0163449 | Retinal dehydrogenase 1 |
| Q96FN9 | -1.6915779 | 0.3690046 | 8.385413 | -4.584165 | 0.0015862 | 0.0163449 | Probable D-tyrosyl-tRNA(Tyr) deacylase 2 |
| Q9UKX3 | 1.6936818 | 0.3854938 | 9.208912 | 4.393538 | 0.0016433 | 0.0168485 | Myosin-13 |
| Q9H1E5 | -1.0100442 | 0.2319493 | 9.385413 | -4.354590 | 0.0016644 | 0.0169760 | Thioredoxin-related transmembrane protein 4 |
| Q9BXV9 | 1.4127212 | 0.3218254 | 9.163202 | 4.389714 | 0.0016725 | 0.0169760 | Uncharacterized protein C14orf142 |
| P80723 | -1.5225978 | 0.3278699 | 7.910668 | -4.643908 | 0.0017078 | 0.0172483 | Brain acid soluble protein 1 |
| A5D6W6 | -2.1874865 | 0.4705266 | 7.868778 | -4.649017 | 0.0017207 | 0.0172842 | Fat storage-inducing transmembrane protein 1 |
| Q8IYI6 | -0.9655844 | 0.2230263 | 9.385413 | -4.329463 | 0.0017284 | 0.0172842 | Exocyst complex component 8 |
| P15924 | 0.9305974 | 0.2148943 | 9.350459 | 4.330490 | 0.0017409 | 0.0173234 | Desmoplakin |
| O15061 | -1.0942259 | 0.2546655 | 9.385413 | -4.296719 | 0.0018158 | 0.0178238 | Synemin |
| Q9UK22 | -1.4762619 | 0.2882849 | 6.372396 | -5.120844 | 0.0018178 | 0.0178238 | F-box only protein 2 |
| P07384 | -0.8789740 | 0.2042255 | 9.333139 | -4.303938 | 0.0018195 | 0.0178238 | Calpain-1 catalytic subunit |
| P52907 | -0.8481991 | 0.1972352 | 9.339311 | -4.300445 | 0.0018263 | 0.0178238 | F-actin-capping protein subunit alpha-1 |
| Q04721 | 1.1127233 | 0.2577419 | 9.201253 | 4.317199 | 0.0018437 | 0.0178982 | Neurogenic locus notch homolog protein 2;Notch 2 extracellular truncation;Notch 2 intracellular domain |
| Q8N142 | 1.0825486 | 0.2509220 | 9.201572 | 4.314284 | 0.0018515 | 0.0178982 | Adenylosuccinate synthetase isozyme 1 |
| O60262 | -1.2355761 | 0.2771111 | 8.388576 | -4.458775 | 0.0018804 | 0.0180907 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-7 |
| Q96A65 | -0.8662506 | 0.2035141 | 9.382448 | -4.256465 | 0.0019311 | 0.0184915 | Exocyst complex component 4 |
| Q9NS69 | -1.0714067 | 0.2476392 | 8.946979 | -4.326483 | 0.0019416 | 0.0185043 | Mitochondrial import receptor subunit TOM22 homolog |
| Q96CX2 | -0.9012081 | 0.2113310 | 9.290787 | -4.264439 | 0.0019510 | 0.0185072 | BTB/POZ domain-containing protein KCTD12 |
| Q6IC98 | -0.9007405 | 0.2081057 | 8.888308 | -4.328283 | 0.0019666 | 0.0185681 | GRAM domain-containing protein 4 |
| P62745 | -1.2315256 | 0.2839409 | 8.788016 | -4.337261 | 0.0019937 | 0.0186338 | Rho-related GTP-binding protein RhoB |
| O00625 | -1.1467230 | 0.2709208 | 9.385413 | -4.232687 | 0.0020006 | 0.0186338 | Pirin |
| P31323 | -0.9308385 | 0.2186352 | 9.229929 | -4.257496 | 0.0020011 | 0.0186338 | cAMP-dependent protein kinase type II-beta regulatory subunit |
| P05455 | -0.8673708 | 0.2037435 | 9.178555 | -4.257171 | 0.0020275 | 0.0187940 | Lupus La protein |
| P61009 | -0.8913019 | 0.2048775 | 8.621704 | -4.350413 | 0.0020468 | 0.0188867 | Signal peptidase complex subunit 3 |
| P62277 | -1.3147062 | 0.3106711 | 9.269754 | -4.231827 | 0.0020596 | 0.0189187 | 40S ribosomal protein S13 |
| P19447 | -1.6209348 | 0.3503038 | 7.385413 | -4.627226 | 0.0020905 | 0.0191159 | TFIIH basal transcription factor complex helicase XPB subunit |
| P13796 | -0.8965848 | 0.2123820 | 9.058132 | -4.221566 | 0.0022024 | 0.0198918 | Plastin-2 |
| P50991 | -1.4847501 | 0.3465048 | 8.685140 | -4.284934 | 0.0022089 | 0.0198918 | T-complex protein 1 subunit delta |
| Q9H1K0 | -1.6569316 | 0.3616836 | 7.385413 | -4.581164 | 0.0022125 | 0.0198918 | Rabenosyn-5 |
| Q9UL18 | -1.3355815 | 0.3024699 | 8.037138 | -4.415585 | 0.0022146 | 0.0198918 | Protein argonaute-1 |
| O75489 | 0.9821574 | 0.2367724 | 9.385413 | 4.148107 | 0.0022758 | 0.0202641 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial |
| Q96C86 | -0.8840200 | 0.2112887 | 9.145534 | -4.183943 | 0.0022805 | 0.0202641 | m7GpppX diphosphatase |
| Q9UBB5 | 1.4054756 | 0.2873545 | 6.385413 | 4.891086 | 0.0022976 | 0.0202641 | Methyl-CpG-binding domain protein 2 |
| Q04760 | 1.3888487 | 0.3250035 | 8.599181 | 4.273334 | 0.0022977 | 0.0202641 | Lactoylglutathione lyase |
| Q12988 | -1.0008329 | 0.2396145 | 9.134204 | -4.176846 | 0.0023111 | 0.0202641 | Heat shock protein beta-3 |
| Q8N5M1 | -1.3322320 | 0.3166085 | 8.937206 | -4.207821 | 0.0023159 | 0.0202641 | ATP synthase mitochondrial F1 complex assembly factor 2 |
| Q6P1N0 | -1.6664832 | 0.3563734 | 6.936107 | -4.676227 | 0.0023268 | 0.0202725 | Coiled-coil and C2 domain-containing protein 1A |
| Q96AG4 | -1.0700845 | 0.2592741 | 9.385413 | -4.127233 | 0.0023497 | 0.0203023 | Leucine-rich repeat-containing protein 59 |
| Q07065 | -0.9428402 | 0.2242953 | 8.903162 | -4.203566 | 0.0023503 | 0.0203023 | Cytoskeleton-associated protein 4 |
| Q9Y4W6 | 0.9288317 | 0.2250637 | 9.237547 | 4.126973 | 0.0024317 | 0.0209168 | AFG3-like protein 2 |
| P00492 | -0.9748591 | 0.2342661 | 8.867949 | -4.161333 | 0.0025227 | 0.0215202 | Hypoxanthine-guanine phosphoribosyltransferase |
| Q8IYU8 | -3.0517638 | 0.6353282 | 6.385413 | -4.803445 | 0.0025231 | 0.0215202 | Calcium uptake protein 2, mitochondrial |
| O75348 | -1.2953207 | 0.3178560 | 9.377764 | -4.075181 | 0.0025496 | 0.0216557 | V-type proton ATPase subunit G 1 |
| Q9UKS6 | 0.9838252 | 0.2380314 | 8.943424 | 4.133174 | 0.0025822 | 0.0218407 | Protein kinase C and casein kinase substrate in neurons protein 3 |
| P50238 | -1.7738249 | 0.4308810 | 8.850185 | -4.116739 | 0.0027058 | 0.0227914 | Cysteine-rich protein 1 |
| P04843 | -1.5463708 | 0.3734394 | 8.660976 | -4.140889 | 0.0027359 | 0.0228771 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 |
| P05543 | -1.2021068 | 0.2678425 | 7.101585 | -4.488111 | 0.0027385 | 0.0228771 | Thyroxine-binding globulin |
| Q96GK7 | -1.2685155 | 0.2966639 | 7.917392 | -4.275936 | 0.0027676 | 0.0230170 | Fumarylacetoacetate hydrolase domain-containing protein 2A |
| P25311 | -1.0760599 | 0.2606542 | 8.673237 | -4.128305 | 0.0027779 | 0.0230170 | Zinc-alpha-2-glycoprotein |
| Q9Y490 | -0.8565056 | 0.2115741 | 9.149741 | -4.048254 | 0.0027957 | 0.0230644 | Talin-1 |
| P58546 | -1.2877434 | 0.3138586 | 8.783206 | -4.102941 | 0.0028064 | 0.0230644 | Myotrophin |
| P53618 | -0.9257503 | 0.2314827 | 9.385413 | -3.999220 | 0.0028622 | 0.0234285 | Coatomer subunit beta |
| P28070 | -0.9313857 | 0.2219436 | 8.111747 | -4.196497 | 0.0029195 | 0.0237512 | Proteasome subunit beta type-4 |
| Q9NNX1 | 3.4364699 | 0.7363292 | 6.385413 | 4.667030 | 0.0029250 | 0.0237512 | Tuftelin |
| Q9BXF6 | -0.9435750 | 0.2370306 | 9.385413 | -3.980816 | 0.0029451 | 0.0238189 | Rab11 family-interacting protein 5 |
| Q14118 | -0.8636209 | 0.2063191 | 8.111452 | -4.185849 | 0.0029629 | 0.0238356 | Dystroglycan;Alpha-dystroglycan;Beta-dystroglycan |
| P21980 | -0.9931919 | 0.2428774 | 8.618924 | -4.089273 | 0.0029798 | 0.0238356 | Protein-glutamine gamma-glutamyltransferase 2 |
| P50552 | -0.9356255 | 0.2329598 | 9.074476 | -4.016253 | 0.0029847 | 0.0238356 | Vasodilator-stimulated phosphoprotein |
| Q92575 | -1.0319247 | 0.2532376 | 8.685218 | -4.074927 | 0.0029941 | 0.0238356 | UBX domain-containing protein 4 |
| Q01581 | -1.6309708 | 0.3718001 | 7.186730 | -4.386687 | 0.0030119 | 0.0238838 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic |
| P31994-2 | -0.8401092 | 0.2094556 | 8.992524 | -4.010918 | 0.0030648 | 0.0242083 | Low affinity immunoglobulin gamma Fc region receptor II-b |
| O75165 | -0.7960642 | 0.1998267 | 9.132663 | -3.983773 | 0.0030957 | 0.0242617 | DnaJ homolog subfamily C member 13 |
| Q9HB40 | -1.4975944 | 0.3192059 | 6.179009 | -4.691625 | 0.0031056 | 0.0242617 | Retinoid-inducible serine carboxypeptidase |
| Q00577 | -1.0160743 | 0.2542684 | 9.027366 | -3.996069 | 0.0031099 | 0.0242617 | Transcriptional activator protein Pur-alpha |
| O15031 | -0.9218013 | 0.2310329 | 9.055589 | -3.989914 | 0.0031194 | 0.0242617 | Plexin-B2 |
| P14625 | -0.9806254 | 0.2486476 | 9.333808 | -3.943837 | 0.0031532 | 0.0244236 | Endoplasmin |
| Q7L4E1 | -1.9857976 | 0.4950092 | 8.847472 | -4.011637 | 0.0031642 | 0.0244236 | Protein FAM73B |
| A6NDG6 | 1.0861342 | 0.2722626 | 8.937374 | 3.989289 | 0.0032057 | 0.0246261 | Phosphoglycolate phosphatase |
| Q15125 | -1.5304848 | 0.3855947 | 9.041599 | -3.969154 | 0.0032291 | 0.0246261 | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase |
| Q2TAY7 | -1.0799883 | 0.2732212 | 9.123335 | -3.952799 | 0.0032522 | 0.0246261 | WD40 repeat-containing protein SMU1;WD40 repeat-containing protein SMU1, N-terminally processed |
| E5RK69 | -1.1331593 | 0.2897068 | 9.385413 | -3.911400 | 0.0032812 | 0.0246261 | Annexin |
| Q14258 | -1.0273754 | 0.2506553 | 8.157903 | -4.098758 | 0.0033032 | 0.0246261 | E3 ubiquitin/ISG15 ligase TRIM25 |
| Q9NS86 | -1.0079781 | 0.2479673 | 8.316103 | -4.064963 | 0.0033269 | 0.0246261 | LanC-like protein 2 |
| Q02818 | -1.1109599 | 0.2603296 | 7.340441 | -4.267513 | 0.0033293 | 0.0246261 | Nucleobindin-1 |
| P43121 | -1.0927918 | 0.2720489 | 8.594930 | -4.016894 | 0.0033306 | 0.0246261 | Cell surface glycoprotein MUC18 |
| Q7KZF4 | -0.8983119 | 0.2302334 | 9.385413 | -3.901744 | 0.0033311 | 0.0246261 | Staphylococcal nuclease domain-containing protein 1 |
| Q9Y2D4 | -1.5301277 | 0.3819079 | 8.653139 | -4.006536 | 0.0033354 | 0.0246261 | Exocyst complex component 6B |
| P01303 | -1.4085709 | 0.3549264 | 8.890312 | -3.968628 | 0.0033419 | 0.0246261 | Pro-neuropeptide Y;Neuropeptide Y;C-flanking peptide of NPY |
| Q9BWJ5 | -1.1951212 | 0.3064683 | 9.385413 | -3.899657 | 0.0033420 | 0.0246261 | Splicing factor 3B subunit 5 |
| P49773 | -0.9630558 | 0.2471701 | 9.385413 | -3.896327 | 0.0033594 | 0.0246261 | Histidine triad nucleotide-binding protein 1 |
| Q15738 | -1.1253828 | 0.2888439 | 9.385413 | -3.896163 | 0.0033603 | 0.0246261 | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating |
| P46063 | -0.8051976 | 0.2054091 | 9.154885 | -3.919971 | 0.0033970 | 0.0248052 | ATP-dependent DNA helicase Q1 |
| Q9HBL0 | 1.0970067 | 0.2708307 | 8.290796 | 4.050526 | 0.0034176 | 0.0248666 | Tensin-1 |
| O95810 | -0.7367195 | 0.1898992 | 9.385413 | -3.879528 | 0.0034489 | 0.0250047 | Serum deprivation-response protein |
| Q8IUX7 | -2.4425715 | 0.5075886 | 5.640778 | -4.812108 | 0.0035035 | 0.0253101 | Adipocyte enhancer-binding protein 1 |
| Q9Y5U8 | -2.4377353 | 0.6307150 | 9.327097 | -3.865035 | 0.0035696 | 0.0256674 | Mitochondrial pyruvate carrier 1 |
| P14555 | -3.1537863 | 0.7281822 | 6.867951 | -4.331040 | 0.0035904 | 0.0256674 | Phospholipase A2, membrane associated |
| Q96PK6 | -0.9222337 | 0.2373986 | 9.148490 | -3.884749 | 0.0035909 | 0.0256674 | RNA-binding protein 14 |
| Q9UHD9 | -0.9017767 | 0.2318045 | 9.043824 | -3.890247 | 0.0036397 | 0.0257977 | Ubiquilin-2 |
| Q96EM0 | -1.2584685 | 0.3144044 | 8.309610 | -4.002706 | 0.0036419 | 0.0257977 | Trans-3-hydroxy-L-proline dehydratase |
| Q9HCN8 | -0.8835166 | 0.2288465 | 9.253672 | -3.860738 | 0.0036473 | 0.0257977 | Stromal cell-derived factor 2-like protein 1 |
| P07360 | -0.9669760 | 0.2462742 | 8.752393 | -3.926420 | 0.0036711 | 0.0258764 | Complement component C8 gamma chain |
| P10109 | 0.8561493 | 0.2221550 | 9.243225 | 3.853837 | 0.0036944 | 0.0259004 | Adrenodoxin, mitochondrial |
| Q9H4A6 | -1.0469452 | 0.2719485 | 9.266844 | -3.849793 | 0.0037001 | 0.0259004 | Golgi phosphoprotein 3 |
| P27695 | -1.4992289 | 0.3923412 | 9.385413 | -3.821237 | 0.0037793 | 0.0263153 | DNA-(apurinic or apyrimidinic site) lyase;DNA-(apurinic or apyrimidinic site) lyase, mitochondrial |
| Q53FA7 | 1.3792842 | 0.3581051 | 9.140607 | 3.851619 | 0.0037853 | 0.0263153 | Quinone oxidoreductase PIG3 |
| Q9NQZ5 | 1.3063706 | 0.3399333 | 9.137333 | 3.843021 | 0.0038384 | 0.0265728 | StAR-related lipid transfer protein 7, mitochondrial |
| Q9UQR1 | -1.5901460 | 0.3824514 | 7.327082 | -4.157773 | 0.0038485 | 0.0265728 | Zinc finger protein 148 |
| Q68DH5 | -1.0294876 | 0.2612363 | 8.365194 | -3.940828 | 0.0039282 | 0.0270311 | LMBR1 domain-containing protein 2 |
| P78406 | -1.7207435 | 0.3887386 | 6.279703 | -4.426479 | 0.0039782 | 0.0270918 | mRNA export factor |
| P62312 | -1.5890450 | 0.4091948 | 8.671810 | -3.883346 | 0.0039851 | 0.0270918 | U6 snRNA-associated Sm-like protein LSm6 |
| P10643 | -1.1553612 | 0.3037929 | 9.241403 | -3.803121 | 0.0039995 | 0.0270918 | Complement component C7 |
| Q9BT09 | -1.2257851 | 0.3203966 | 9.058668 | -3.825837 | 0.0040051 | 0.0270918 | Protein canopy homolog 3 |
| Q6PCE3 | -1.3823379 | 0.3363631 | 7.385413 | -4.109659 | 0.0040266 | 0.0270918 | Glucose 1,6-bisphosphate synthase |
| Q9BVC6 | -1.1861599 | 0.3132913 | 9.341936 | -3.786124 | 0.0040278 | 0.0270918 | Transmembrane protein 109 |
| P00488 | -1.1640817 | 0.3067565 | 9.268393 | -3.794807 | 0.0040304 | 0.0270918 | Coagulation factor XIII A chain |
| Q3ZCW2 | -1.4639580 | 0.3351832 | 6.410138 | -4.367635 | 0.0040470 | 0.0271133 | Galectin-related protein |
| Q96KP1 | -1.0715375 | 0.2583008 | 7.188491 | -4.148409 | 0.0040611 | 0.0271186 | Exocyst complex component 2 |
| P61925 | 1.3829061 | 0.3292482 | 6.953044 | 4.200194 | 0.0040956 | 0.0271960 | cAMP-dependent protein kinase inhibitor alpha |
| Q9UNN8 | -1.2626877 | 0.3237195 | 8.433185 | -3.900561 | 0.0040995 | 0.0271960 | Endothelial protein C receptor |
| O00299 | -0.9207980 | 0.2441733 | 9.349959 | -3.771084 | 0.0041180 | 0.0272298 | Chloride intracellular channel protein 1 |
| Q8IVD9 | -0.9866036 | 0.2486289 | 7.962021 | -3.968177 | 0.0041691 | 0.0274783 | NudC domain-containing protein 3 |
| O15121 | -1.4079273 | 0.3728328 | 9.216343 | -3.776297 | 0.0041910 | 0.0275334 | Sphingolipid delta(4)-desaturase DES1 |
| P04207 | -1.4228021 | 0.3725405 | 8.861844 | -3.819188 | 0.0042135 | 0.0275916 | Ig kappa chain V-III region CLL |
| P24311 | 1.0701358 | 0.2851747 | 9.295713 | 3.752563 | 0.0042839 | 0.0279625 | Cytochrome c oxidase subunit 7B, mitochondrial |
| O75663 | -1.7379175 | 0.4581589 | 8.939622 | -3.793263 | 0.0043137 | 0.0280667 | TIP41-like protein |
| Q8NBF2 | -0.8250417 | 0.2215677 | 9.379760 | -3.723655 | 0.0044137 | 0.0285351 | NHL repeat-containing protein 2 |
| P61020 | -0.8382949 | 0.2205335 | 8.768454 | -3.801213 | 0.0044149 | 0.0285351 | Ras-related protein Rab-5B |
| P60903 | -0.8876449 | 0.2381638 | 9.333841 | -3.727035 | 0.0044279 | 0.0285351 | Protein S100-A10 |
| P41208 | -1.1263858 | 0.2999947 | 9.075440 | -3.754686 | 0.0044552 | 0.0286207 | Centrin-2 |
| P20042 | -0.8887367 | 0.2370873 | 9.065935 | -3.748564 | 0.0045061 | 0.0287075 | Eukaryotic translation initiation factor 2 subunit 2 |
| P27169 | -1.0504292 | 0.2769234 | 8.718029 | -3.793212 | 0.0045159 | 0.0287075 | Serum paraoxonase/arylesterase 1 |
| Q08357 | -1.2684736 | 0.3315172 | 8.463053 | -3.826267 | 0.0045409 | 0.0287075 | Sodium-dependent phosphate transporter 2 |
| Q16204 | -0.8049274 | 0.2138365 | 8.902922 | -3.764218 | 0.0045433 | 0.0287075 | Coiled-coil domain-containing protein 6 |
| Q8WUM0 | -0.7917468 | 0.2110572 | 8.990261 | -3.751338 | 0.0045540 | 0.0287075 | Nuclear pore complex protein Nup133 |
| P00918 | -0.9510512 | 0.2547612 | 9.125781 | -3.733109 | 0.0045623 | 0.0287075 | Carbonic anhydrase 2 |
| Q96MM6 | 1.7864789 | 0.4212053 | 6.459250 | 4.241349 | 0.0045992 | 0.0287075 | Heat shock 70 kDa protein 12B |
| P31689 | -1.4924000 | 0.3848993 | 8.072563 | -3.877378 | 0.0046114 | 0.0287075 | DnaJ homolog subfamily A member 1 |
| P49756 | -0.8931713 | 0.2418433 | 9.385413 | -3.693182 | 0.0046276 | 0.0287075 | RNA-binding protein 25 |
| Q5JUQ0 | 2.7926112 | 0.6712936 | 6.726504 | 4.160044 | 0.0046289 | 0.0287075 | Protein FAM78A |
| P01611 | -1.3131481 | 0.3557280 | 9.385413 | -3.691439 | 0.0046405 | 0.0287075 | Ig kappa chain V-I region Wes |
| P32119 | -0.9674030 | 0.2451869 | 7.636769 | -3.945574 | 0.0046754 | 0.0287075 | Peroxiredoxin-2 |
| P62330 | -0.8152583 | 0.2199163 | 9.196457 | -3.707130 | 0.0046879 | 0.0287075 | ADP-ribosylation factor 6 |
| O75643 | -0.7616501 | 0.2067379 | 9.385413 | -3.684133 | 0.0046947 | 0.0287075 | U5 small nuclear ribonucleoprotein 200 kDa helicase |
| P17050 | -0.9853240 | 0.2652090 | 9.121362 | -3.715273 | 0.0046949 | 0.0287075 | Alpha-N-acetylgalactosaminidase |
| P31949 | -0.9787100 | 0.2647594 | 9.277157 | -3.696602 | 0.0046950 | 0.0287075 | Protein S100-A11;Protein S100-A11, N-terminally processed |
| Q9Y646 | -0.7162308 | 0.1940218 | 9.210822 | -3.691497 | 0.0047915 | 0.0291484 | Carboxypeptidase Q |
| Q1KMD3 | -0.9795111 | 0.2640085 | 9.051770 | -3.710149 | 0.0047958 | 0.0291484 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 |
| Q6ZVF9 | -1.5028535 | 0.3517719 | 6.221356 | -4.272239 | 0.0048341 | 0.0292737 | G protein-regulated inducer of neurite outgrowth 3 |
| Q9BU61 | -1.2998963 | 0.3549162 | 9.385413 | -3.662545 | 0.0048588 | 0.0292737 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3 |
| Q9Y5U9 | -0.9729708 | 0.2545202 | 8.183105 | -3.822764 | 0.0048597 | 0.0292737 | Immediate early response 3-interacting protein 1 |
| Q8TCJ2 | -0.9004011 | 0.2429306 | 8.907652 | -3.706413 | 0.0049600 | 0.0297891 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit STT3B |
| Q15493 | -1.6077032 | 0.4360378 | 9.046599 | -3.687073 | 0.0049758 | 0.0297963 | Regucalcin |
| Q9UBS4 | -1.7446965 | 0.4698184 | 8.803956 | -3.713555 | 0.0050071 | 0.0298953 | DnaJ homolog subfamily B member 11 |
| Q9BVG4 | -1.4704698 | 0.4039345 | 9.385413 | -3.640367 | 0.0050337 | 0.0299660 | Protein PBDC1 |
| O15144 | -1.0444705 | 0.2843210 | 9.070426 | -3.673560 | 0.0050589 | 0.0299983 | Actin-related protein 2/3 complex subunit 2 |
| Q9BXR6 | -1.3501527 | 0.3714481 | 9.382448 | -3.634835 | 0.0050810 | 0.0299983 | Complement factor H-related protein 5 |
| O43290 | -0.7923809 | 0.2180343 | 9.385413 | -3.634203 | 0.0050835 | 0.0299983 | U4/U6.U5 tri-snRNP-associated protein 1 |
| Q04837 | -1.2562850 | 0.3379135 | 8.636848 | -3.717771 | 0.0051451 | 0.0302738 | Single-stranded DNA-binding protein, mitochondrial |
| O00567 | -1.5854812 | 0.4377198 | 9.376799 | -3.622138 | 0.0051902 | 0.0304511 | Nucleolar protein 56 |
| Q04941 | -1.3384979 | 0.3673776 | 9.127049 | -3.643385 | 0.0052480 | 0.0307016 | Proteolipid protein 2 |
| Q6P1L8 | 0.6869586 | 0.1895585 | 9.186165 | 3.623993 | 0.0053529 | 0.0312208 | 39S ribosomal protein L14, mitochondrial |
| P21281 | -1.3587904 | 0.3769568 | 9.344431 | -3.604631 | 0.0053675 | 0.0312208 | V-type proton ATPase subunit B, brain isoform |
| Q8TDB6 | -0.9160565 | 0.2529440 | 9.153547 | -3.621578 | 0.0054049 | 0.0312627 | E3 ubiquitin-protein ligase DTX3L |
| P56199 | -0.9018380 | 0.2448251 | 8.647866 | -3.683600 | 0.0054055 | 0.0312627 | Integrin alpha-1 |
| Q8IWU2 | -0.7746135 | 0.2080306 | 8.224391 | -3.723556 | 0.0055613 | 0.0319924 | Serine/threonine-protein kinase LMTK2 |
| Q9Y623 | -2.1252883 | 0.5700777 | 8.189261 | -3.728068 | 0.0055670 | 0.0319924 | Myosin-4 |
| P46777 | -1.0470686 | 0.2879004 | 8.852405 | -3.636912 | 0.0055790 | 0.0319924 | 60S ribosomal protein L5 |
| Q96PE7 | -1.1531979 | 0.3230087 | 9.385413 | -3.570176 | 0.0056319 | 0.0321622 | Methylmalonyl-CoA epimerase, mitochondrial |
| Q6ICB0 | -2.0977670 | 0.5236139 | 6.696096 | -4.006324 | 0.0056403 | 0.0321622 | Desumoylating isopeptidase 1 |
| P30050 | -1.1234823 | 0.3143996 | 9.261423 | -3.573421 | 0.0057230 | 0.0325424 | 60S ribosomal protein L12 |
| P08754 | -1.0611744 | 0.2984515 | 9.385413 | -3.555601 | 0.0057651 | 0.0326906 | Guanine nucleotide-binding protein G(k) subunit alpha |
| P11021 | -0.7515167 | 0.2089700 | 8.970360 | -3.596290 | 0.0058129 | 0.0328607 | 78 kDa glucose-regulated protein |
| P56192 | -1.0557634 | 0.2923102 | 8.813819 | -3.611792 | 0.0058414 | 0.0328607 | Methionine–tRNA ligase, cytoplasmic |
| Q53GG5 | 1.0003784 | 0.2774742 | 8.865410 | 3.605302 | 0.0058437 | 0.0328607 | PDZ and LIM domain protein 3 |
| Q8NDY3 | 1.2195639 | 0.3410277 | 9.101142 | 3.576143 | 0.0058602 | 0.0328626 | [Protein ADP-ribosylarginine] hydrolase-like protein 1 |
| O75533 | -0.7695176 | 0.2164213 | 9.256606 | -3.555646 | 0.0058921 | 0.0329503 | Splicing factor 3B subunit 1 |
| P01699 | -1.8429363 | 0.4533786 | 6.314916 | -4.064895 | 0.0059388 | 0.0331204 | Ig lambda chain V-I region VOR |
| Q14019 | -2.4182957 | 0.5985491 | 6.385413 | -4.040263 | 0.0059733 | 0.0331901 | Coactosin-like protein |
| Q9Y5J7 | -1.4119681 | 0.3997182 | 9.385413 | -3.532409 | 0.0059840 | 0.0331901 | Mitochondrial import inner membrane translocase subunit Tim9 |
| P23284 | -0.7777143 | 0.2189746 | 9.127890 | -3.551619 | 0.0060629 | 0.0335357 | Peptidyl-prolyl cis-trans isomerase B |
| Q9Y5Z7 | -1.8906077 | 0.4616964 | 6.110612 | -4.094916 | 0.0061500 | 0.0339251 | Host cell factor 2 |
| Q9HAV7 | 0.8784715 | 0.2455573 | 8.806708 | 3.577460 | 0.0061670 | 0.0339271 | GrpE protein homolog 1, mitochondrial |
| P08670 | -1.0385574 | 0.2890644 | 8.643328 | -3.592824 | 0.0062100 | 0.0340714 | Vimentin |
| Q9H9B4 | -0.7835938 | 0.2188802 | 8.717170 | -3.580012 | 0.0062456 | 0.0341738 | Sideroflexin-1 |
| Q5T447 | -1.0539901 | 0.2946359 | 8.718056 | -3.577263 | 0.0062709 | 0.0342204 | E3 ubiquitin-protein ligase HECTD3 |
| P05060 | -2.3172970 | 0.6115268 | 7.305570 | -3.789363 | 0.0062887 | 0.0342252 | Secretogranin-1;PE-11;GAWK peptide;CCB peptide |
| P14174 | -0.9252890 | 0.2597430 | 8.795118 | -3.562324 | 0.0063263 | 0.0343378 | Macrophage migration inhibitory factor |
| P00742 | -1.3823666 | 0.3377126 | 6.021781 | -4.093323 | 0.0063561 | 0.0343429 | Coagulation factor X;Factor X light chain;Factor X heavy chain;Activated factor Xa heavy chain |
| P05062 | -2.0691367 | 0.5491873 | 7.376614 | -3.767634 | 0.0063611 | 0.0343429 | Fructose-bisphosphate aldolase B |
| P01597 | -2.2245190 | 0.6295868 | 8.995469 | -3.533300 | 0.0063859 | 0.0343855 | Ig kappa chain V-I region DEE |
| Q9UK41 | -1.5840168 | 0.4525342 | 9.249370 | -3.500325 | 0.0064436 | 0.0346045 | Vacuolar protein sorting-associated protein 28 homolog |
| O14807 | -1.0071551 | 0.2870529 | 9.142433 | -3.508604 | 0.0064740 | 0.0346760 | Ras-related protein M-Ras |
| Q9GZY4 | -1.6315958 | 0.4423737 | 7.703311 | -3.688275 | 0.0065660 | 0.0349087 | Cytochrome c oxidase assembly factor 1 homolog |
| Q13011 | 0.8125674 | 0.2281934 | 8.603557 | 3.560872 | 0.0065690 | 0.0349087 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| A4D2B0 | -1.7771506 | 0.4954576 | 8.385413 | -3.586888 | 0.0065854 | 0.0349087 | Metallo-beta-lactamase domain-containing protein 1 |
| P27797 | -0.8785716 | 0.2529576 | 9.382448 | -3.473197 | 0.0065862 | 0.0349087 | Calreticulin |
| Q9HAN9 | 1.2290381 | 0.3277463 | 7.306422 | 3.749968 | 0.0066297 | 0.0350474 | Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1 |
| P02749 | -1.0092251 | 0.2887067 | 9.025558 | -3.495676 | 0.0067400 | 0.0355380 | Beta-2-glycoprotein 1 |
| Q14011 | -1.6557884 | 0.4613486 | 8.231467 | -3.589018 | 0.0067678 | 0.0355925 | Cold-inducible RNA-binding protein |
| P62191 | -0.9985121 | 0.2875705 | 9.176854 | -3.472234 | 0.0068202 | 0.0357754 | 26S protease regulatory subunit 4 |
| Q8NBJ5 | -1.0369389 | 0.2922708 | 8.473099 | -3.547871 | 0.0068666 | 0.0357787 | Procollagen galactosyltransferase 1 |
| P16455 | -0.8416366 | 0.2363383 | 8.365652 | -3.561152 | 0.0068706 | 0.0357787 | Methylated-DNA–protein-cysteine methyltransferase |
| Q12907 | -1.6608858 | 0.4654093 | 8.305704 | -3.568657 | 0.0068737 | 0.0357787 | Vesicular integral-membrane protein VIP36 |
| Q9UMR3 | -1.7064633 | 0.4609378 | 7.385413 | -3.702155 | 0.0069386 | 0.0360239 | T-box transcription factor TBX20 |
| Q5VIR6-4 | -0.9082357 | 0.2626254 | 9.130015 | -3.458294 | 0.0070272 | 0.0363239 | Vacuolar protein sorting-associated protein 53 homolog |
| P06396 | -1.1602520 | 0.3284971 | 8.474042 | -3.532001 | 0.0070322 | 0.0363239 | Gelsolin |
| Q6Y288 | -1.0921514 | 0.3170900 | 9.229532 | -3.444295 | 0.0070709 | 0.0364313 | Beta-1,3-glucosyltransferase |
| P98160 | -0.7123685 | 0.2057393 | 9.004949 | -3.462481 | 0.0071269 | 0.0366270 | Basement membrane-specific heparan sulfate proteoglycan core protein;Endorepellin;LG3 peptide |
| Q9HCJ6 | -0.9011150 | 0.2633086 | 9.382448 | -3.422277 | 0.0071515 | 0.0366605 | Synaptic vesicle membrane protein VAT-1 homolog-like |
| O43707 | -1.3824302 | 0.3940398 | 8.555847 | -3.508352 | 0.0071799 | 0.0367132 | Alpha-actinin-4 |
| Q13425 | -0.7846763 | 0.2289447 | 9.241730 | -3.427362 | 0.0072514 | 0.0369860 | Beta-2-syntrophin |
| Q9UBI9 | -1.9787149 | 0.5456575 | 7.613412 | -3.626295 | 0.0073101 | 0.0371672 | Headcase protein homolog |
| Q13636 | -1.9278246 | 0.4828349 | 5.941397 | -3.992720 | 0.0073236 | 0.0371672 | Ras-related protein Rab-31 |
| O43592 | -0.8599780 | 0.2513274 | 9.208263 | -3.421744 | 0.0073560 | 0.0372388 | Exportin-T |
| P43686 | -1.3071386 | 0.3842049 | 9.385413 | -3.402192 | 0.0073848 | 0.0372913 | 26S protease regulatory subunit 6B |
| O75190-3 | 1.2139008 | 0.3579316 | 9.385413 | 3.391433 | 0.0075148 | 0.0378540 | DnaJ homolog subfamily B member 6 |
| Q9NX08 | -0.7560262 | 0.2199925 | 8.886277 | -3.436601 | 0.0075720 | 0.0379630 | COMM domain-containing protein 8 |
| Q96CN7 | -1.0345528 | 0.3035605 | 9.151255 | -3.408061 | 0.0075873 | 0.0379630 | Isochorismatase domain-containing protein 1 |
| O60568 | -1.0499114 | 0.3101574 | 9.385413 | -3.385092 | 0.0075926 | 0.0379630 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| O60927 | -0.9982244 | 0.2669526 | 6.797580 | -3.739332 | 0.0076677 | 0.0381466 | Protein phosphatase 1 regulatory subunit 11 |
| Q9H2J4 | -0.8634690 | 0.2556745 | 9.385413 | -3.377220 | 0.0076903 | 0.0381466 | Phosducin-like protein 3 |
| Q8NFQ8 | -0.7967626 | 0.2241386 | 7.843418 | -3.554776 | 0.0077012 | 0.0381466 | Torsin-1A-interacting protein 2 |
| P15848 | 0.9063750 | 0.2622189 | 8.604816 | 3.456558 | 0.0077045 | 0.0381466 | Arylsulfatase B |
| P30101 | -0.7466004 | 0.2200397 | 9.129332 | -3.393025 | 0.0077988 | 0.0385196 | Protein disulfide-isomerase A3 |
| P51692 | -1.7183021 | 0.5058169 | 9.071768 | -3.397083 | 0.0078195 | 0.0385280 | Signal transducer and activator of transcription 5B |
| Q96LD4 | -2.0992980 | 0.5326176 | 5.906119 | -3.941473 | 0.0078555 | 0.0386116 | Tripartite motif-containing protein 47 |
| Q96G03 | -0.8867606 | 0.2633690 | 9.330409 | -3.366989 | 0.0078842 | 0.0386591 | Phosphoglucomutase-2 |
| P05387 | -0.7376313 | 0.2197628 | 9.385413 | -3.356488 | 0.0079539 | 0.0389072 | 60S acidic ribosomal protein P2 |
| Q08AG7 | -1.2342110 | 0.3426996 | 7.362325 | -3.601437 | 0.0080096 | 0.0390851 | Mitotic-spindle organizing protein 1 |
| Q9Y678 | -0.6484637 | 0.1937556 | 9.385413 | -3.346813 | 0.0080802 | 0.0393350 | Coatomer subunit gamma-1 |
| P29992 | -1.0188326 | 0.2947365 | 8.303781 | -3.456757 | 0.0081310 | 0.0394877 | Guanine nucleotide-binding protein subunit alpha-11 |
| Q9BX97 | -1.3844164 | 0.3942853 | 7.837641 | -3.511205 | 0.0082107 | 0.0397799 | Plasmalemma vesicle-associated protein |
| P16083 | -1.0262131 | 0.3000391 | 8.385413 | -3.420264 | 0.0084662 | 0.0409198 | Ribosyldihydronicotinamide dehydrogenase [quinone] |
| P03915 | -1.1707120 | 0.3448778 | 8.492156 | -3.394570 | 0.0086415 | 0.0416682 | NADH-ubiquinone oxidoreductase chain 5 |
| Q13825 | 1.4155209 | 0.4286271 | 9.385413 | 3.302453 | 0.0086860 | 0.0417125 | Methylglutaconyl-CoA hydratase, mitochondrial |
| P55735 | -0.8920338 | 0.2672114 | 8.993847 | -3.338307 | 0.0086918 | 0.0417125 | Protein SEC13 homolog |
| Q13541 | 0.9219952 | 0.2801292 | 9.385413 | 3.291322 | 0.0088453 | 0.0423489 | Eukaryotic translation initiation factor 4E-binding protein 1 |
| Q9BSH5 | -0.8053114 | 0.2432271 | 9.081702 | -3.310944 | 0.0089588 | 0.0427913 | Haloacid dehalogenase-like hydrolase domain-containing protein 3 |
| O95159 | -1.8090106 | 0.5289166 | 8.060595 | -3.420219 | 0.0089825 | 0.0428040 | Zinc finger protein-like 1 |
| P61225 | -0.7353323 | 0.2254094 | 9.385413 | -3.262208 | 0.0092764 | 0.0440350 | Ras-related protein Rap-2b |
| O75436 | -1.0001875 | 0.2995476 | 8.574417 | -3.338993 | 0.0092842 | 0.0440350 | Vacuolar protein sorting-associated protein 26A |
| P22748 | 1.3439356 | 0.3988665 | 8.227398 | 3.369388 | 0.0094030 | 0.0444942 | Carbonic anhydrase 4 |
| O43920 | 0.7871398 | 0.2413857 | 9.266721 | 3.260921 | 0.0094526 | 0.0446249 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 |
| B0YJ81 | -0.8197164 | 0.2437757 | 8.223227 | -3.362585 | 0.0095069 | 0.0447771 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 1 |
| P61026 | -0.8525514 | 0.2469341 | 7.519029 | -3.452546 | 0.0095312 | 0.0447877 | Ras-related protein Rab-10 |
| Q5VUM1 | 0.7593853 | 0.2321285 | 9.045228 | 3.271401 | 0.0095967 | 0.0449156 | Succinate dehydrogenase assembly factor 4, mitochondrial |
| Q8N392 | 2.3525552 | 0.6309863 | 6.037587 | 3.728378 | 0.0096451 | 0.0449156 | Rho GTPase-activating protein 18 |
| Q8N129 | -0.9589743 | 0.2874231 | 8.368916 | -3.336456 | 0.0096458 | 0.0449156 | Protein canopy homolog 4 |
| P22897 | -1.2570252 | 0.3866597 | 9.236964 | -3.250986 | 0.0096469 | 0.0449156 | Macrophage mannose receptor 1 |
| I3L505 | 1.1736484 | 0.3548842 | 8.571534 | 3.307130 | 0.0097586 | 0.0453318 | Acyl carrier protein |
| Q99459 | -0.8211683 | 0.2419414 | 7.796908 | -3.394079 | 0.0098104 | 0.0454681 | Cell division cycle 5-like protein |
| Q13061 | -2.4676439 | 0.7427573 | 8.281135 | -3.322275 | 0.0100042 | 0.0462607 | Triadin |
| Q9UBQ0 | -1.1062507 | 0.3422745 | 9.158961 | -3.232057 | 0.0100576 | 0.0464023 | Vacuolar protein sorting-associated protein 29 |
| P01621 | -1.2058630 | 0.3710231 | 8.927861 | -3.250102 | 0.0101023 | 0.0465026 | Ig kappa chain V-III region NG9 |
| O14656 | -1.8276459 | 0.5541078 | 8.385413 | -3.298358 | 0.0101964 | 0.0468296 | Torsin-1A |
| P41743 | -1.1383453 | 0.3178404 | 6.465679 | -3.581500 | 0.0102399 | 0.0469233 | Protein kinase C iota type |
| P09467 | -1.3548161 | 0.3766861 | 6.379503 | -3.596671 | 0.0102775 | 0.0469893 | Fructose-1,6-bisphosphatase 1 |
| P04430 | -1.0635292 | 0.3314758 | 9.250027 | -3.208467 | 0.0103183 | 0.0470701 | Ig kappa chain V-I region BAN |
| Q9H4G4 | -1.8410577 | 0.5694674 | 8.904758 | -3.232947 | 0.0104175 | 0.0474158 | Golgi-associated plant pathogenesis-related protein 1 |
| P35754 | 0.7959584 | 0.2488883 | 9.268764 | 3.198055 | 0.0104680 | 0.0475244 | Glutaredoxin-1 |
| Q08211 | -0.7008973 | 0.2199769 | 9.382448 | -3.186232 | 0.0105108 | 0.0475244 | ATP-dependent RNA helicase A |
| Q6UXG3 | 1.6056385 | 0.5021363 | 9.243547 | 3.197615 | 0.0105116 | 0.0475244 | CMRF35-like molecule 9 |
| Q9HD45 | -0.9322808 | 0.2920818 | 9.281520 | -3.191848 | 0.0105563 | 0.0476206 | Transmembrane 9 superfamily member 3 |
| O00487 | -0.7979909 | 0.2500603 | 9.242344 | -3.191194 | 0.0106238 | 0.0478188 | 26S proteasome non-ATPase regulatory subunit 14 |
| Q86WH2 | -1.4748731 | 0.3898450 | 5.524605 | -3.783230 | 0.0106687 | 0.0479148 | Ras association domain-containing protein 3 |
| P08648 | -0.9830382 | 0.3010637 | 8.373691 | -3.265216 | 0.0107486 | 0.0481672 | Integrin alpha-5;Integrin alpha-5 heavy chain;Integrin alpha-5 light chain |
| P07196 | -3.1755881 | 0.8558310 | 5.733074 | -3.710532 | 0.0108167 | 0.0483656 | Neurofilament light polypeptide |
| P00156 | -1.7108654 | 0.5393991 | 9.325104 | -3.171799 | 0.0108447 | 0.0483803 | Cytochrome b |
| P34947 | -1.4297892 | 0.4514007 | 9.363114 | -3.167450 | 0.0108677 | 0.0483803 | G protein-coupled receptor kinase 5 |
| Q6B0K9 | -1.3489774 | 0.4080821 | 7.853752 | -3.305652 | 0.0110508 | 0.0490876 | Hemoglobin subunit mu |
| Q92616 | -0.5950798 | 0.1874991 | 9.031070 | -3.173775 | 0.0112487 | 0.0498578 | Translational activator GCN1 |
| Q92805 | 1.3236880 | 0.4166069 | 8.947665 | 3.177307 | 0.0113157 | 0.0499740 | Golgin subfamily A member 1 |
| Q14165 | -1.0392026 | 0.3299855 | 9.277055 | -3.149237 | 0.0113242 | 0.0499740 | Malectin |
| P51665 | -0.7776248 | 0.2465259 | 9.194991 | -3.154333 | 0.0113529 | 0.0499924 | 26S proteasome non-ATPase regulatory subunit 7 |
4.6 Interaction
4.6.1 Volcano-plot
volcanoInt <- ggplot(rowData(pe[["proteinRobust"]])$"locationR:tissueV",
aes(x = logFC, y = -log10(pval), color = adjPval < 0.05)) +
geom_point(cex = 2.5) +
scale_color_manual(values = alpha(c("black", "red"), 0.5)) + theme_minimal()
volcanoInt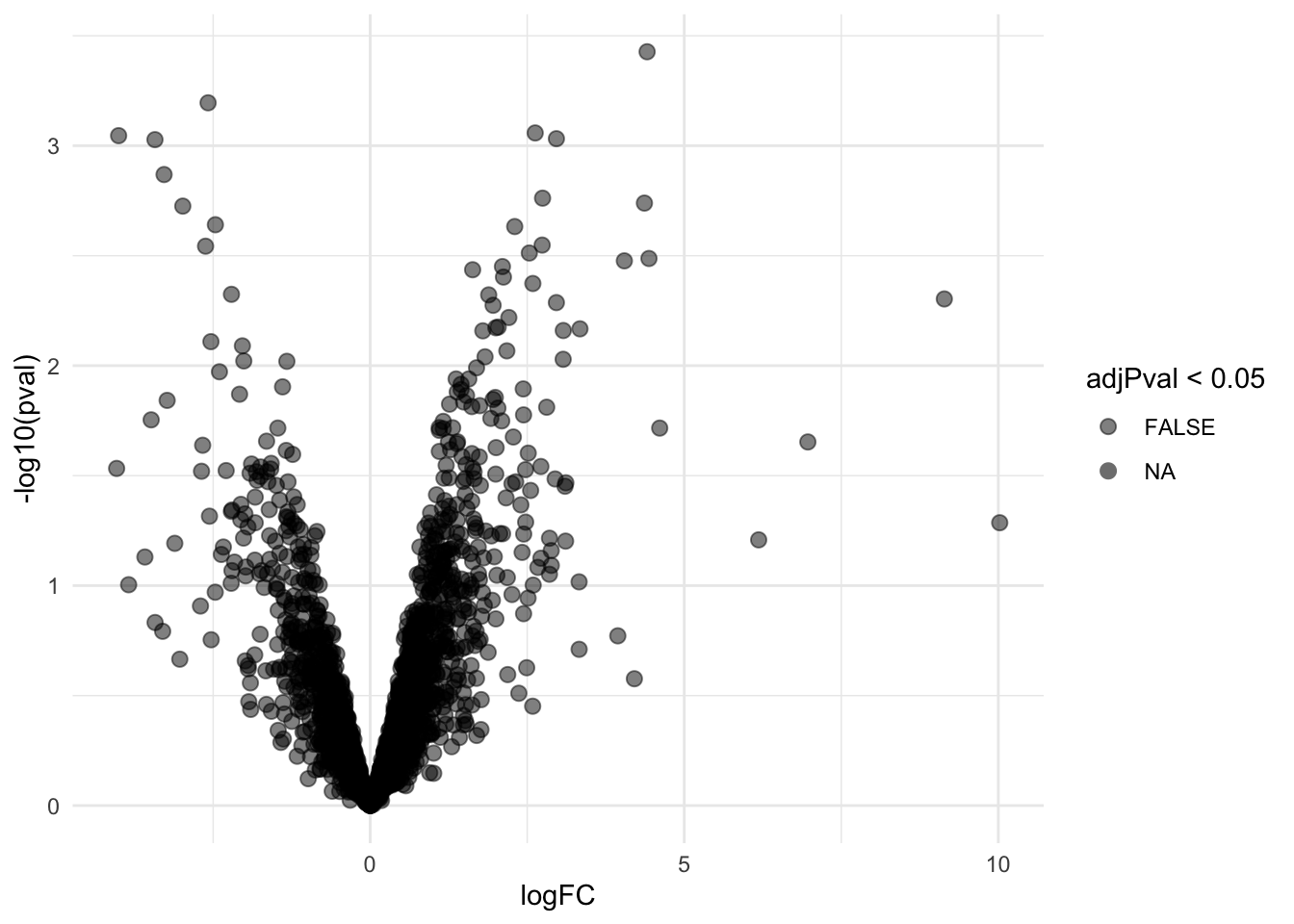
4.6.2 Heatmap
There were no genes significant at the 5% FDR level. We return the top 25 genes.
sigNamesInt <- rowData(pe[["proteinRobust"]])$"locationR:tissueV" %>%
rownames_to_column("proteinRobust") %>%
filter(adjPval<0.05) %>%
pull(proteinRobust)
hlp <- order((rowData(pe[["proteinRobust"]])$"locationR:tissueV")[,"adjPval"])[1:25]
heatmap(assay(pe[["proteinRobust"]])[hlp, ])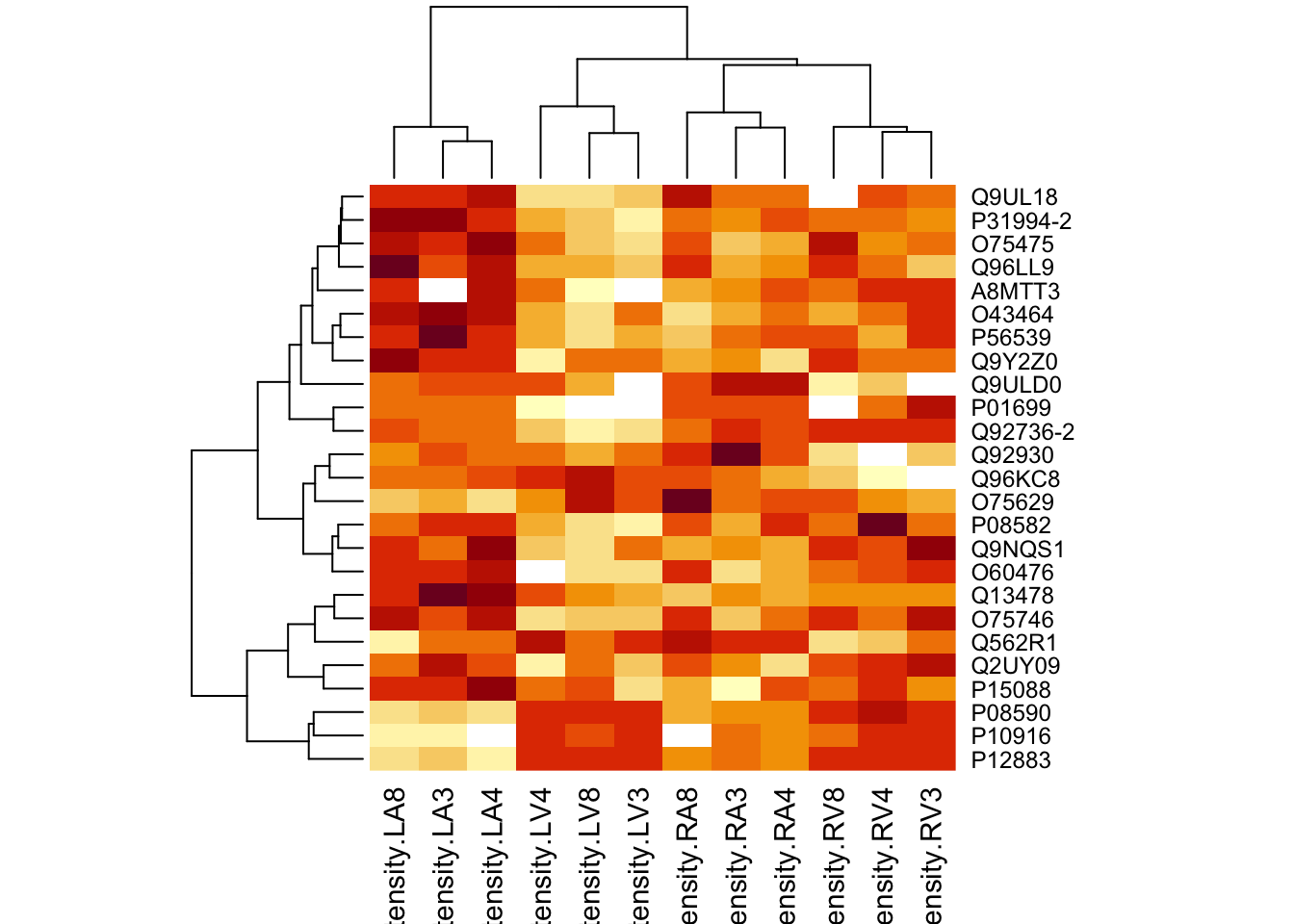
There are 0 proteins significantly differentially expressed at the 5% FDR level.
rowData(pe[["proteinRobust"]])$"locationR:tissueV" %>%
cbind(.,rowData(pe[["proteinRobust"]])$Protein.names) %>%
na.exclude %>%
filter(adjPval<0.05) %>%
arrange(pval)5 Large difference in number of proteins that are returned
Note, that much more proteins are returned significant for average contrast (\(\log_2 FC_{V-A}\)) as compared to contrast for assessing the fold change between ventriculum and atrium left and right. The power for the average contrast is larger than for the contrast left or right because the log2 FC can be estimated with higher precision.
For none of the proteins the interaction was significant (change in log2 FC between ventriculum and atrium in the right vs the left heart region). The power for the interaction is typically low.
5.1 Reason
Part of variance covariance matrix of model parameters due to design:
Variance of contrasts (diagonal elements) due to design
## tissueV tissueV + locationR:tissueV
## 0.6666667 0.6666667
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.3333333 1.3333333## tissueV tissueV + locationR:tissueV
## 0.8164966 0.8164966
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.5773503 1.1547005So it is clear that the standard error of the log2 FC left and right is the same. That of the average contrast is a factor \(\sqrt{2}\) smaller! Indeed, we use double the number of samples to estimate it!
## tissueV + 0.5 * locationR:tissueV
## 0.5## tissueV + 0.5 * locationR:tissueV
## 0.7071068## [1] 0.7071068The standard error of the interaction is a factor \(\sqrt{2}\) larger than that of the main effects!
## locationR:tissueV
## 2## locationR:tissueV
## 1.414214## [1] 1.4142145.2 Msqrob
This is not the case for the standard errors of protein 2???
Because msqrob is using robust regression to assess DE!
pe %>%
colData %>%
as_tibble %>%
mutate(w=getModel(rowData(pe[["proteinRobust"]])$msqrobModels[[2]])$w)For protein 2 the samples at the left and right side have different weights!
5.2.1 Part of standard error due to design:
covUnscaledRobust <- solve(
t(X) %*%
diag(
getModel(rowData(pe[["proteinRobust"]])$msqrobModels[[2]])$w) %*% X)
varContrastsRobust <- t(L)%*%covUnscaledRobust%*%L %>%
diag
varContrastsRobust## tissueV tissueV + locationR:tissueV
## 0.7187863 0.8704210
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.3950893 1.5980575## tissueV tissueV + locationR:tissueV
## 0.8478127 0.9329636
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.6285613 1.26414305.2.2 Standard errors Contrasts
## tissueV tissueV + locationR:tissueV
## 0.5601628 0.6164233
## tissueV + 0.5 * locationR:tissueV locationR:tissueV
## 0.4153001 0.83523875.3 Stagewise testing
5.3.1 Omnibus test
5.3.2 Heatmap significant proteins Omnibus test
We first select the names of the proteins that were declared signficant.
sigNamesOmnibus <- rowData(pe[["proteinRobust"]])$omnibusTest %>%
rownames_to_column("proteinRobust") %>%
filter(adjPval<0.05) %>%
pull(proteinRobust)
heatmap(assay(pe[["proteinRobust"]])[sigNamesOmnibus, ])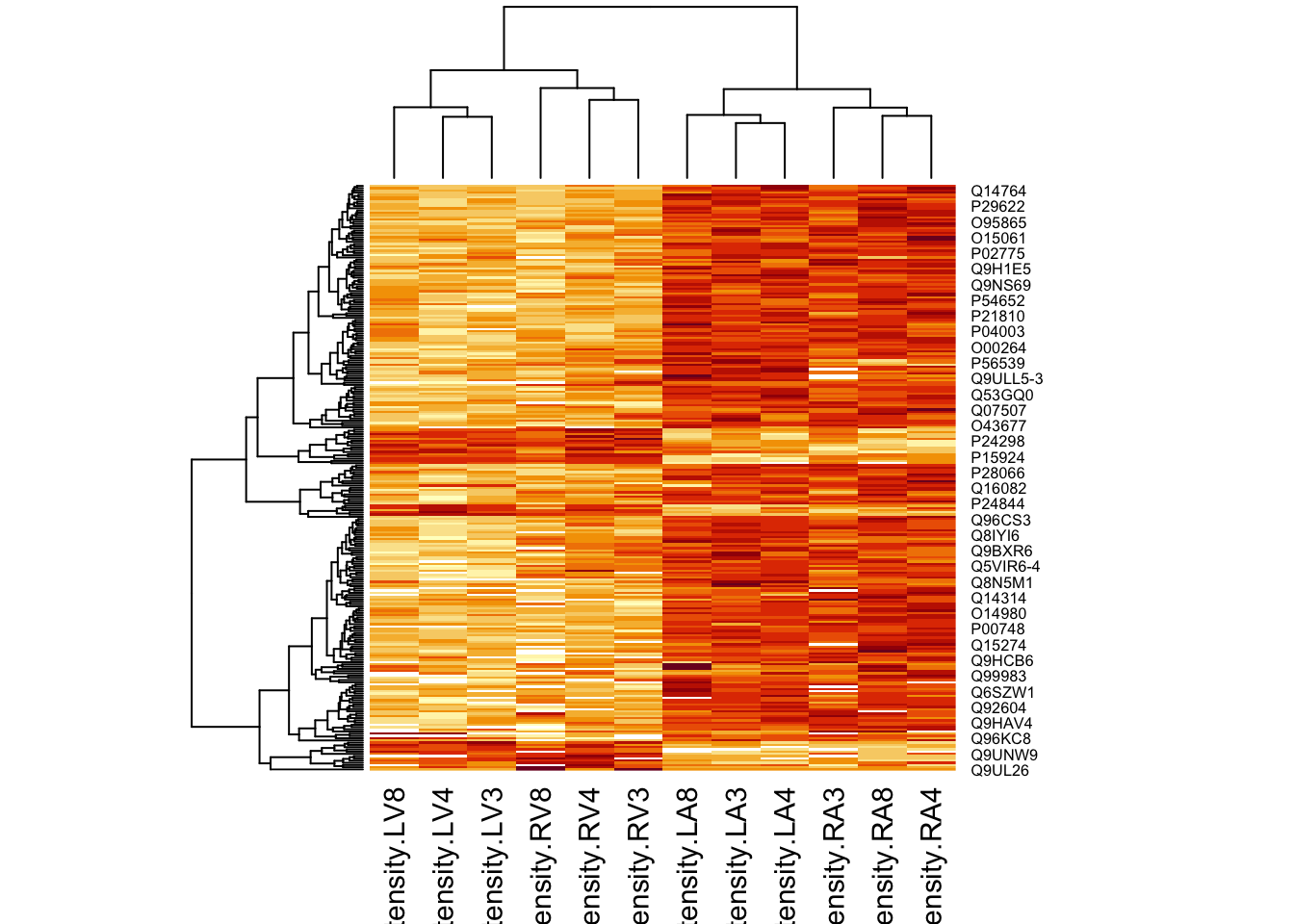
There are 262 proteins significantly differentially expressed at the 5% FDR level.
rowData(pe[["proteinRobust"]])$omnibusTest %>%
cbind(.,rowData(pe[["proteinRobust"]])$Protein.names) %>%
na.exclude %>%
filter(adjPval<0.05) %>%
arrange(pval) %>%
knitr::kable(.)| tissueV | tissueV…locationR.tissueV | tissueV…0.5…locationR.tissueV | locationR.tissueV | df1 | df2 | Fstat | pval | adjPval | rowData(pe[[“proteinRobust”]])$Protein.names | |
|---|---|---|---|---|---|---|---|---|---|---|
| P08590 | 7.5669453 | 5.3580772 | 6.4625112 | -2.2088680 | 2 | 9.117912 | 241.941690 | 0.0000000 | 0.0000255 | Myosin light chain 3 |
| P12883 | 4.3331248 | 1.7523890 | 3.0427569 | -2.5807358 | 2 | 9.255194 | 82.809688 | 0.0000012 | 0.0012591 | Myosin-7 |
| P10916 | 6.9086744 | 2.9035913 | 4.9061329 | -4.0050831 | 2 | 7.385413 | 107.580783 | 0.0000035 | 0.0023247 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| O75368 | -2.2903758 | -1.8440754 | -2.0672256 | 0.4463004 | 2 | 9.251939 | 59.046905 | 0.0000054 | 0.0023247 | SH3 domain-binding glutamic acid-rich-like protein |
| P06858 | 1.7952437 | 3.3292464 | 2.5622451 | 1.5340027 | 2 | 9.218266 | 56.616101 | 0.0000066 | 0.0023247 | Lipoprotein lipase |
| Q6UWY5 | -3.2532286 | -2.4696667 | -2.8614477 | 0.7835619 | 2 | 9.170135 | 55.962919 | 0.0000073 | 0.0023247 | Olfactomedin-like protein 1 |
| P46821 | -2.1828217 | -1.6229630 | -1.9028923 | 0.5598587 | 2 | 9.385413 | 52.504004 | 0.0000080 | 0.0023247 | Microtubule-associated protein 1B;MAP1B heavy chain;MAP1 light chain LC1 |
| P29622 | -2.1148699 | -1.8621552 | -1.9885126 | 0.2527147 | 2 | 9.385413 | 44.363771 | 0.0000165 | 0.0039435 | Kallistatin |
| P05546 | -1.9156713 | -1.7501838 | -1.8329275 | 0.1654876 | 2 | 9.198639 | 45.181005 | 0.0000175 | 0.0039435 | Heparin cofactor 2 |
| P21810 | -3.3802538 | -2.9942069 | -3.1872304 | 0.3860470 | 2 | 8.304055 | 51.174076 | 0.0000214 | 0.0042345 | Biglycan |
| P35442 | -2.3342598 | -3.2070907 | -2.7706752 | -0.8728309 | 2 | 8.385413 | 49.249094 | 0.0000232 | 0.0042345 | Thrombospondin-2 |
| P13533 | -4.3139246 | -4.0839508 | -4.1989377 | 0.2299738 | 2 | 9.115028 | 42.042011 | 0.0000250 | 0.0042345 | Myosin-6 |
| Q8N474 | -3.3046470 | -2.1617235 | -2.7331853 | 1.1429234 | 2 | 7.847514 | 52.144823 | 0.0000294 | 0.0045872 | Secreted frizzled-related protein 1 |
| Q06828 | -4.2091891 | -4.1354763 | -4.1723327 | 0.0737128 | 2 | 9.003303 | 39.385102 | 0.0000353 | 0.0051216 | Fibromodulin |
| P02452 | -3.2261931 | -2.0070368 | -2.6166150 | 1.2191563 | 2 | 8.367424 | 42.481732 | 0.0000415 | 0.0056136 | Collagen alpha-1(I) chain |
| O95865 | -2.1589316 | -0.9959370 | -1.5774343 | 1.1629946 | 2 | 9.385413 | 34.272459 | 0.0000486 | 0.0057994 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 |
| O94875-10 | 2.3212380 | 1.3818264 | 1.8515322 | -0.9394116 | 2 | 8.958355 | 36.659139 | 0.0000486 | 0.0057994 | Sorbin and SH3 domain-containing protein 2 |
| Q16647 | -2.7876976 | -1.5342676 | -2.1609826 | 1.2534300 | 2 | 9.156725 | 34.947347 | 0.0000517 | 0.0058198 | Prostacyclin synthase |
| Q9UGT4 | -2.2836150 | -2.4665348 | -2.3750749 | -0.1829198 | 2 | 9.193722 | 34.308942 | 0.0000545 | 0.0058198 | Sushi domain-containing protein 2 |
| P02776 | -1.7487175 | -2.8417114 | -2.2952145 | -1.0929939 | 2 | 7.718405 | 43.279869 | 0.0000639 | 0.0062978 | Platelet factor 4;Platelet factor 4, short form |
| A6NMZ7 | -2.2673287 | -3.0659585 | -2.6666436 | -0.7986298 | 2 | 9.385413 | 31.905851 | 0.0000652 | 0.0062978 | Collagen alpha-6(VI) chain |
| Q14764 | -1.6259758 | -1.6162557 | -1.6211158 | 0.0097201 | 2 | 9.385413 | 31.218467 | 0.0000712 | 0.0065710 | Major vault protein |
| Q15113 | -2.7550209 | -1.9885085 | -2.3717647 | 0.7665124 | 2 | 9.188121 | 31.518143 | 0.0000770 | 0.0067277 | Procollagen C-endopeptidase enhancer 1 |
| Q9ULL5-3 | -3.3863077 | -1.2106027 | -2.2984552 | 2.1757051 | 2 | 7.893242 | 39.193814 | 0.0000796 | 0.0067277 | Proline-rich protein 12 |
| Q9P2B2 | -2.0475095 | -2.0452709 | -2.0463902 | 0.0022386 | 2 | 9.385413 | 30.078364 | 0.0000829 | 0.0067277 | Prostaglandin F2 receptor negative regulator |
| P14854 | 2.3006633 | 1.0658853 | 1.6832743 | -1.2347780 | 2 | 8.346841 | 33.671446 | 0.0001009 | 0.0075831 | Cytochrome c oxidase subunit 6B1 |
| P18428 | -2.0025775 | -1.7014100 | -1.8519938 | 0.3011674 | 2 | 9.013977 | 29.954553 | 0.0001043 | 0.0075831 | Lipopolysaccharide-binding protein |
| P54652 | -1.5859216 | -2.7252456 | -2.1555836 | -1.1393240 | 2 | 9.007166 | 29.940008 | 0.0001049 | 0.0075831 | Heat shock-related 70 kDa protein 2 |
| P24844 | -2.5518015 | -1.7836148 | -2.1677081 | 0.7681867 | 2 | 9.330669 | 28.358116 | 0.0001083 | 0.0075831 | Myosin regulatory light polypeptide 9 |
| P51888 | -3.1995487 | -1.9468369 | -2.5731928 | 1.2527118 | 2 | 6.959924 | 42.594973 | 0.0001247 | 0.0081957 | Prolargin |
| Q9BW30 | -2.7386840 | -2.3263164 | -2.5325002 | 0.4123676 | 2 | 9.049210 | 28.443735 | 0.0001252 | 0.0081957 | Tubulin polymerization-promoting protein family member 3 |
| O43677 | -2.7580901 | -2.9654848 | -2.8617874 | -0.2073947 | 2 | 7.933870 | 33.631479 | 0.0001335 | 0.0082984 | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial |
| P00748 | -2.0030563 | -1.9489960 | -1.9760261 | 0.0540602 | 2 | 9.091376 | 27.735769 | 0.0001349 | 0.0082984 | Coagulation factor XII;Coagulation factor XIIa heavy chain;Beta-factor XIIa part 1;Coagulation factor XIIa light chain |
| P48681 | -1.3300808 | -1.4933277 | -1.4117043 | -0.1632468 | 2 | 9.321612 | 26.157695 | 0.0001501 | 0.0089644 | Nestin |
| P02775 | -1.9521262 | -2.1690290 | -2.0605776 | -0.2169029 | 2 | 9.385413 | 25.639727 | 0.0001573 | 0.0090542 | Platelet basic protein;Connective tissue-activating peptide III;TC-2;Connective tissue-activating peptide III(1-81);Beta-thromboglobulin;Neutrophil-activating peptide 2(74);Neutrophil-activating peptide 2(73);Neutrophil-activating peptide 2;TC-1;Neutrophil-activating peptide 2(1-66);Neutrophil-activating peptide 2(1-63) |
| P08294 | -2.7564583 | -1.8981910 | -2.3273246 | 0.8582673 | 2 | 7.979491 | 31.192375 | 0.0001691 | 0.0090542 | Extracellular superoxide dismutase [Cu-Zn] |
| P23083 | -4.2742932 | -2.2755749 | -3.2749341 | 1.9987183 | 2 | 7.385413 | 35.088072 | 0.0001693 | 0.0090542 | Ig heavy chain V-I region V35 |
| P36955 | -2.3236197 | -1.0657353 | -1.6946775 | 1.2578843 | 2 | 8.456893 | 28.729923 | 0.0001695 | 0.0090542 | Pigment epithelium-derived factor |
| P02743 | -2.2737781 | -1.6607108 | -1.9672445 | 0.6130673 | 2 | 9.385413 | 24.776129 | 0.0001801 | 0.0093742 | Serum amyloid P-component;Serum amyloid P-component(1-203) |
| O00180 | -4.0967217 | -4.7553461 | -4.4260339 | -0.6586245 | 2 | 8.385413 | 28.272422 | 0.0001875 | 0.0095158 | Potassium channel subfamily K member 1 |
| P51884 | -2.4022338 | -1.4533109 | -1.9277723 | 0.9489230 | 2 | 7.936878 | 29.702993 | 0.0002064 | 0.0099663 | Lumican |
| P00325 | -2.1088138 | -0.8991435 | -1.5039787 | 1.2096703 | 2 | 9.141096 | 24.684486 | 0.0002066 | 0.0099663 | Alcohol dehydrogenase 1B |
| Q5NDL2 | -2.3376295 | -2.7045789 | -2.5211042 | -0.3669494 | 2 | 8.686198 | 25.916946 | 0.0002180 | 0.0099663 | EGF domain-specific O-linked N-acetylglucosamine transferase |
| Q92604 | -2.2702815 | -1.9257953 | -2.0980384 | 0.3444862 | 2 | 9.345516 | 23.715791 | 0.0002181 | 0.0099663 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 |
| Q53GQ0 | -2.4308529 | -1.2156054 | -1.8232291 | 1.2152475 | 2 | 9.334954 | 23.651824 | 0.0002215 | 0.0099663 | Very-long-chain 3-oxoacyl-CoA reductase |
| P60468 | -1.5229242 | -2.4541896 | -1.9885569 | -0.9312654 | 2 | 8.241407 | 27.348175 | 0.0002300 | 0.0099663 | Protein transport protein Sec61 subunit beta |
| P07451 | -1.8415388 | -0.7406077 | -1.2910732 | 1.1009311 | 2 | 9.385413 | 23.260186 | 0.0002307 | 0.0099663 | Carbonic anhydrase 3 |
| Q92508 | 4.0710006 | 2.9423812 | 3.5066909 | -1.1286194 | 2 | 6.385413 | 40.291134 | 0.0002393 | 0.0101201 | Piezo-type mechanosensitive ion channel component 1 |
| Q6PCB0 | -1.3297806 | -2.3007919 | -1.8152862 | -0.9710113 | 2 | 9.198450 | 23.100847 | 0.0002591 | 0.0107360 | von Willebrand factor A domain-containing protein 1 |
| P28066 | -1.1096114 | -2.1707758 | -1.6401936 | -1.0611644 | 2 | 8.500773 | 25.051701 | 0.0002730 | 0.0108002 | Proteasome subunit alpha type-5 |
| Q9UBG0 | -2.5762099 | -0.7476767 | -1.6619433 | 1.8285332 | 2 | 9.131671 | 22.977636 | 0.0002731 | 0.0108002 | C-type mannose receptor 2 |
| P30405 | -1.5402735 | -1.8282016 | -1.6842375 | -0.2879280 | 2 | 9.385413 | 22.200005 | 0.0002767 | 0.0108002 | Peptidyl-prolyl cis-trans isomerase F, mitochondrial |
| P01031 | -1.3223783 | -1.6526586 | -1.4875185 | -0.3302802 | 2 | 8.183937 | 25.866283 | 0.0002898 | 0.0111012 | Complement C5;Complement C5 beta chain;Complement C5 alpha chain;C5a anaphylatoxin;Complement C5 alpha chain |
| P05997 | -3.0688637 | -1.9991154 | -2.5339895 | 1.0697482 | 2 | 8.864434 | 23.034598 | 0.0003082 | 0.0114681 | Collagen alpha-2(V) chain |
| Q9ULD0 | -0.5232010 | -3.5070276 | -2.0151143 | -2.9838266 | 2 | 6.128068 | 39.648177 | 0.0003119 | 0.0114681 | 2-oxoglutarate dehydrogenase-like, mitochondrial |
| Q15327 | -1.3545314 | -2.2217826 | -1.7881570 | -0.8672512 | 2 | 9.280457 | 21.682263 | 0.0003179 | 0.0114681 | Ankyrin repeat domain-containing protein 1 |
| Q8TBP6 | -1.8932428 | -1.5272808 | -1.7102618 | 0.3659619 | 2 | 8.851865 | 22.802350 | 0.0003220 | 0.0114681 | Solute carrier family 25 member 40 |
| Q14195-2 | -2.5272781 | -1.9904875 | -2.2588828 | 0.5367907 | 2 | 9.262403 | 21.354010 | 0.0003397 | 0.0115202 | Dihydropyrimidinase-related protein 3 |
| P35625 | -1.9848027 | -3.9792035 | -2.9820031 | -1.9944008 | 2 | 7.962134 | 25.589243 | 0.0003412 | 0.0115202 | Metalloproteinase inhibitor 3 |
| O14980 | -1.2303520 | -1.0484463 | -1.1393991 | 0.1819057 | 2 | 9.382448 | 20.935701 | 0.0003473 | 0.0115202 | Exportin-1 |
| Q8WWA0 | -5.8069752 | -3.0892056 | -4.4480904 | 2.7177697 | 2 | 8.244689 | 24.246612 | 0.0003521 | 0.0115202 | Intelectin-1 |
| P46060 | -1.9706423 | -1.5112752 | -1.7409587 | 0.4593671 | 2 | 8.385413 | 23.700667 | 0.0003543 | 0.0115202 | Ran GTPase-activating protein 1 |
| P36021 | -3.1383009 | -1.9362318 | -2.5372663 | 1.2020690 | 2 | 8.385413 | 23.640542 | 0.0003575 | 0.0115202 | Monocarboxylate transporter 8 |
| Q69YU5 | 1.7637858 | 3.2208428 | 2.4923143 | 1.4570570 | 2 | 8.841525 | 21.866238 | 0.0003778 | 0.0119831 | Uncharacterized protein C12orf73 |
| O95980 | -2.5524206 | -1.3836422 | -1.9680314 | 1.1687784 | 2 | 7.980660 | 24.630656 | 0.0003851 | 0.0120269 | Reversion-inducing cysteine-rich protein with Kazal motifs |
| Q7L4S7 | -1.7474498 | -2.5477609 | -2.1476054 | -0.8003111 | 2 | 7.157555 | 27.778526 | 0.0004233 | 0.0128614 | Protein ARMCX6 |
| Q8TBQ9 | -2.7618057 | -0.7242810 | -1.7430434 | 2.0375247 | 2 | 8.170120 | 23.247359 | 0.0004245 | 0.0128614 | Protein kish-A |
| P04004 | -1.6339729 | -1.9329355 | -1.7834542 | -0.2989626 | 2 | 8.981078 | 20.612102 | 0.0004402 | 0.0128719 | Vitronectin;Vitronectin V65 subunit;Vitronectin V10 subunit;Somatomedin-B |
| P12110 | -2.1364562 | -1.2366997 | -1.6865779 | 0.8997564 | 2 | 8.535409 | 21.831782 | 0.0004403 | 0.0128719 | Collagen alpha-2(VI) chain |
| P41240 | -1.5691477 | -1.1536153 | -1.3613815 | 0.4155324 | 2 | 8.922337 | 20.715046 | 0.0004439 | 0.0128719 | Tyrosine-protein kinase CSK |
| O15230 | -1.4150113 | -1.3512669 | -1.3831391 | 0.0637444 | 2 | 9.044210 | 19.855982 | 0.0004913 | 0.0138534 | Laminin subunit alpha-5 |
| Q9NZ01 | -2.3460403 | -1.3540953 | -1.8500678 | 0.9919450 | 2 | 8.691035 | 20.740126 | 0.0004914 | 0.0138534 | Very-long-chain enoyl-CoA reductase |
| Q07954 | -1.6748321 | -0.9719344 | -1.3233833 | 0.7028977 | 2 | 9.385413 | 18.940512 | 0.0005073 | 0.0140821 | Prolow-density lipoprotein receptor-related protein 1;Low-density lipoprotein receptor-related protein 1 85 kDa subunit;Low-density lipoprotein receptor-related protein 1 515 kDa subunit;Low-density lipoprotein receptor-related protein 1 intracellular domain |
| Q9BXN1 | -2.6709234 | -1.9091316 | -2.2900275 | 0.7617918 | 2 | 8.937487 | 19.871226 | 0.0005133 | 0.0140821 | Asporin |
| Q15274 | -1.9282879 | -2.1785858 | -2.0534369 | -0.2502979 | 2 | 7.757883 | 23.240643 | 0.0005296 | 0.0141879 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] |
| Q6YN16 | 1.5271671 | 1.5467735 | 1.5369703 | 0.0196064 | 2 | 9.222955 | 19.046286 | 0.0005312 | 0.0141879 | Hydroxysteroid dehydrogenase-like protein 2 |
| P04083 | -1.5245384 | -1.1749000 | -1.3497192 | 0.3496384 | 2 | 9.385413 | 18.571596 | 0.0005462 | 0.0143987 | Annexin A1 |
| P04003 | -1.4790527 | -1.3920940 | -1.4355734 | 0.0869587 | 2 | 9.090898 | 19.007793 | 0.0005655 | 0.0147162 | C4b-binding protein alpha chain |
| Q9UL18 | -2.6023120 | -0.0688510 | -1.3355815 | 2.5334610 | 2 | 8.037138 | 21.366792 | 0.0006069 | 0.0154288 | Protein argonaute-1 |
| O60760 | -3.5000288 | -1.0631593 | -2.2815940 | 2.4368695 | 2 | 8.385413 | 20.152153 | 0.0006268 | 0.0154288 | Hematopoietic prostaglandin D synthase |
| Q9Y6X5 | -1.4114347 | -1.5449603 | -1.4781975 | -0.1335256 | 2 | 8.750753 | 19.161143 | 0.0006350 | 0.0154288 | Bis(5-adenosyl)-triphosphatase ENPP4 |
| P23142 | -1.8527598 | -2.8190443 | -2.3359021 | -0.9662845 | 2 | 7.590129 | 22.566639 | 0.0006390 | 0.0154288 | Fibulin-1 |
| Q92736-2 | -3.3307404 | 1.0781855 | -1.1262774 | 4.4089259 | 2 | 8.908498 | 18.724478 | 0.0006447 | 0.0154288 | Ryanodine receptor 2 |
| Q96LL9 | -2.4266405 | -0.1255693 | -1.2761049 | 2.3010712 | 2 | 8.842792 | 18.847708 | 0.0006474 | 0.0154288 | DnaJ homolog subfamily C member 30 |
| O75828 | -1.6417394 | -1.3689557 | -1.5053475 | 0.2727838 | 2 | 9.385413 | 17.702832 | 0.0006530 | 0.0154288 | Carbonyl reductase [NADPH] 3 |
| P04196 | -1.9171807 | -1.4596228 | -1.6884017 | 0.4575579 | 2 | 7.683140 | 22.070959 | 0.0006536 | 0.0154288 | Histidine-rich glycoprotein |
| P30711 | -1.2535537 | -2.3235254 | -1.7885395 | -1.0699717 | 2 | 9.018645 | 18.192306 | 0.0006834 | 0.0159469 | Glutathione S-transferase theta-1 |
| P23434 | 1.1268218 | 1.4139379 | 1.2703798 | 0.2871161 | 2 | 9.385413 | 17.425133 | 0.0006923 | 0.0159707 | Glycine cleavage system H protein, mitochondrial |
| Q5JPH6 | 3.5060169 | 3.2437535 | 3.3748852 | -0.2622634 | 2 | 6.212234 | 28.881640 | 0.0007149 | 0.0163056 | Probable glutamate–tRNA ligase, mitochondrial |
| Q92681 | -2.1530541 | -1.4997046 | -1.8263794 | 0.6533496 | 2 | 8.133279 | 19.834425 | 0.0007448 | 0.0167504 | Regulatory solute carrier protein family 1 member 1 |
| Q9BZH6 | 1.9311553 | 3.4204862 | 2.6758207 | 1.4893309 | 2 | 8.367230 | 19.171225 | 0.0007509 | 0.0167504 | WD repeat-containing protein 11 |
| P49207 | -1.5348292 | -1.2156675 | -1.3752484 | 0.3191617 | 2 | 8.970773 | 17.586091 | 0.0007870 | 0.0173652 | 60S ribosomal protein L34 |
| P07357 | -1.1549122 | -1.2564148 | -1.2056635 | -0.1015026 | 2 | 9.290174 | 16.879989 | 0.0008065 | 0.0174685 | Complement component C8 alpha chain |
| P04275 | -1.0540996 | -1.3623248 | -1.2082122 | -0.3082252 | 2 | 8.991784 | 17.410957 | 0.0008089 | 0.0174685 | von Willebrand factor;von Willebrand antigen 2 |
| P48163 | -0.8742508 | -2.9539287 | -1.9140897 | -2.0796779 | 2 | 7.960353 | 19.749073 | 0.0008201 | 0.0175233 | NADP-dependent malic enzyme |
| Q9NRG4 | 2.1576737 | 2.5530787 | 2.3553762 | 0.3954050 | 2 | 8.385413 | 18.346713 | 0.0008659 | 0.0182138 | N-lysine methyltransferase SMYD2 |
| Q9BUF5 | -1.9343777 | -1.6205004 | -1.7774391 | 0.3138773 | 2 | 9.217423 | 16.653562 | 0.0008703 | 0.0182138 | Tubulin beta-6 chain |
| P50453 | -1.0571443 | -1.4224494 | -1.2397969 | -0.3653051 | 2 | 9.052869 | 16.851253 | 0.0008877 | 0.0183877 | Serpin B9 |
| P40261 | -2.3067271 | -0.5611559 | -1.4339415 | 1.7455712 | 2 | 9.340024 | 16.214682 | 0.0009165 | 0.0186174 | Nicotinamide N-methyltransferase |
| O14967 | -2.3931306 | -0.8808598 | -1.6369952 | 1.5122708 | 2 | 8.430438 | 17.938253 | 0.0009171 | 0.0186174 | Calmegin |
| Q9HCB6 | -1.8088797 | -2.4163742 | -2.1126269 | -0.6074945 | 2 | 8.648303 | 17.394332 | 0.0009313 | 0.0186367 | Spondin-1 |
| Q86WV6 | -1.0645731 | -1.3366433 | -1.2006082 | -0.2720702 | 2 | 9.239827 | 16.281293 | 0.0009364 | 0.0186367 | Stimulator of interferon genes protein |
| P01024 | -1.1482973 | -1.1510164 | -1.1496569 | -0.0027191 | 2 | 9.197920 | 16.269998 | 0.0009533 | 0.0187877 | Complement C3;Complement C3 beta chain;C3-beta-c;Complement C3 alpha chain;C3a anaphylatoxin;Acylation stimulating protein;Complement C3b alpha chain;Complement C3c alpha chain fragment 1;Complement C3dg fragment;Complement C3g fragment;Complement C3d fragment;Complement C3f fragment;Complement C3c alpha chain fragment 2 |
| Q8WZA9 | -1.5022008 | -0.7123621 | -1.1072814 | 0.7898387 | 2 | 9.101344 | 16.199917 | 0.0010030 | 0.0192980 | Immunity-related GTPase family Q protein |
| Q8WY22 | -1.8394420 | -1.2560002 | -1.5477211 | 0.5834418 | 2 | 8.385413 | 17.535888 | 0.0010096 | 0.0192980 | BRI3-binding protein |
| O00264 | -1.9992421 | -0.8347285 | -1.4169853 | 1.1645136 | 2 | 9.079390 | 16.165900 | 0.0010187 | 0.0192980 | Membrane-associated progesterone receptor component 1 |
| P49458 | -2.3208229 | -1.5193157 | -1.9200693 | 0.8015072 | 2 | 7.120983 | 20.998598 | 0.0010323 | 0.0192980 | Signal recognition particle 9 kDa protein |
| Q00G26 | 0.9705589 | 1.9366811 | 1.4536200 | 0.9661222 | 2 | 7.788675 | 18.796138 | 0.0010453 | 0.0192980 | Perilipin-5 |
| P24298 | 1.3010640 | 1.9631743 | 1.6321192 | 0.6621103 | 2 | 8.623072 | 16.855736 | 0.0010485 | 0.0192980 | Alanine aminotransferase 1 |
| P12814 | -1.7261945 | -2.1924895 | -1.9593420 | -0.4662950 | 2 | 8.144626 | 17.867453 | 0.0010509 | 0.0192980 | Alpha-actinin-1 |
| P62760 | -1.9296447 | -1.8525435 | -1.8910941 | 0.0771012 | 2 | 9.120970 | 15.908955 | 0.0010620 | 0.0192980 | Visinin-like protein 1 |
| Q92621 | -1.7555813 | -0.7290578 | -1.2423196 | 1.0265235 | 2 | 9.385413 | 15.486770 | 0.0010647 | 0.0192980 | Nuclear pore complex protein Nup205 |
| P14550 | -1.3940797 | -1.2147931 | -1.3044364 | 0.1792867 | 2 | 8.301450 | 17.330401 | 0.0010881 | 0.0195469 | Alcohol dehydrogenase [NADP(+)] |
| Q9UHG2 | -1.5781315 | -3.3052829 | -2.4417072 | -1.7271515 | 2 | 8.796297 | 16.293511 | 0.0011019 | 0.0195842 | ProSAAS;KEP;Big SAAS;Little SAAS;Big PEN-LEN;PEN;Little LEN;Big LEN |
| Q9UQ35 | -1.3675109 | -1.1283860 | -1.2479485 | 0.2391249 | 2 | 9.385413 | 15.310560 | 0.0011095 | 0.0195842 | Serine/arginine repetitive matrix protein 2 |
| Q9UNW9 | 4.0970526 | 2.5592726 | 3.3281626 | -1.5377800 | 2 | 9.054127 | 15.777880 | 0.0011203 | 0.0196049 | RNA-binding protein Nova-2 |
| P02747 | -2.6809142 | -1.0489855 | -1.8649499 | 1.6319287 | 2 | 7.138784 | 20.155201 | 0.0011582 | 0.0200961 | Complement C1q subcomponent subunit C |
| O94919 | -1.1585816 | -1.1506677 | -1.1546247 | 0.0079139 | 2 | 9.248102 | 15.281924 | 0.0011709 | 0.0201441 | Endonuclease domain-containing 1 protein |
| Q9UBB5 | 2.6733014 | 0.1376498 | 1.4054756 | -2.5356516 | 2 | 6.385413 | 22.969050 | 0.0012118 | 0.0206722 | Methyl-CpG-binding domain protein 2 |
| Q9Y3B4 | -1.2545628 | -1.3433144 | -1.2989386 | -0.0887516 | 2 | 9.218746 | 15.129931 | 0.0012255 | 0.0207013 | Splicing factor 3B subunit 6 |
| Q92930 | -0.1821330 | -2.8031266 | -1.4926298 | -2.6209936 | 2 | 7.597170 | 18.350597 | 0.0012339 | 0.0207013 | Ras-related protein Rab-8B |
| Q09161 | 1.6000882 | 2.4230607 | 2.0115745 | 0.8229726 | 2 | 6.358037 | 22.719676 | 0.0012705 | 0.0210567 | Nuclear cap-binding protein subunit 1 |
| Q9ULC3 | -1.6748081 | -1.2521086 | -1.4634583 | 0.4226994 | 2 | 8.669265 | 15.832512 | 0.0012759 | 0.0210567 | Ras-related protein Rab-23 |
| Q5M9N0 | -3.5370655 | -2.0324007 | -2.7847331 | 1.5046647 | 2 | 8.350040 | 16.314342 | 0.0013050 | 0.0213638 | Coiled-coil domain-containing protein 158 |
| Q6SZW1 | -2.5636873 | -1.5366583 | -2.0501728 | 1.0270290 | 2 | 7.385413 | 18.512262 | 0.0013274 | 0.0215565 | Sterile alpha and TIR motif-containing protein 1 |
| Q53GG5-2 | -2.9233739 | -1.9547211 | -2.4390475 | 0.9686528 | 2 | 8.429675 | 16.001178 | 0.0013491 | 0.0215717 | PDZ and LIM domain protein 3 |
| P31994-2 | -1.6555661 | -0.0246523 | -0.8401092 | 1.6309138 | 2 | 8.992524 | 15.052398 | 0.0013496 | 0.0215717 | Low affinity immunoglobulin gamma Fc region receptor II-b |
| P08603 | -0.8156387 | -1.4932560 | -1.1544473 | -0.6776173 | 2 | 9.385413 | 14.422402 | 0.0013730 | 0.0217748 | Complement factor H |
| Q96FN9 | -0.7351941 | -2.6479617 | -1.6915779 | -1.9127675 | 2 | 8.385413 | 15.887535 | 0.0014054 | 0.0221164 | Probable D-tyrosyl-tRNA(Tyr) deacylase 2 |
| P11586 | 0.5240804 | 2.0119956 | 1.2680380 | 1.4879152 | 2 | 7.881516 | 16.870189 | 0.0014189 | 0.0221574 | C-1-tetrahydrofolate synthase, cytoplasmic;Methylenetetrahydrofolate dehydrogenase;Methenyltetrahydrofolate cyclohydrolase;Formyltetrahydrofolate synthetase;C-1-tetrahydrofolate synthase, cytoplasmic, N-terminally processed |
| Q9H1K0 | -1.0302150 | -2.2836483 | -1.6569316 | -1.2534333 | 2 | 7.385413 | 18.059570 | 0.0014323 | 0.0221947 | Rabenosyn-5 |
| Q9BUH6 | -0.7048278 | -1.5164145 | -1.1106212 | -0.8115867 | 2 | 9.115985 | 14.547929 | 0.0014559 | 0.0223900 | Protein PAXX |
| Q53T59 | -1.8521243 | -1.8154181 | -1.8337712 | 0.0367062 | 2 | 7.876061 | 16.521863 | 0.0015200 | 0.0230978 | HCLS1-binding protein 3 |
| P25940 | -1.6803001 | -1.5733256 | -1.6268129 | 0.1069744 | 2 | 9.043783 | 14.456074 | 0.0015247 | 0.0230978 | Collagen alpha-3(V) chain |
| Q9NY15 | -2.1774225 | -0.7897346 | -1.4835786 | 1.3876880 | 2 | 9.336638 | 14.011059 | 0.0015443 | 0.0232216 | Stabilin-1 |
| Q8NAT1 | -1.4164507 | -0.3174685 | -0.8669596 | 1.0989822 | 2 | 9.385413 | 13.914217 | 0.0015581 | 0.0232449 | Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 |
| O43464 | -1.9007206 | 0.7261872 | -0.5872667 | 2.6269078 | 2 | 9.072346 | 14.297299 | 0.0015687 | 0.0232449 | Serine protease HTRA2, mitochondrial |
| P07585 | -1.9272925 | -2.3418627 | -2.1345776 | -0.4145702 | 2 | 9.163968 | 14.130035 | 0.0015853 | 0.0233194 | Decorin |
| Q99983 | -2.4961356 | -3.5746196 | -3.0353776 | -1.0784839 | 2 | 6.904704 | 18.806830 | 0.0016058 | 0.0234514 | Osteomodulin |
| Q9BS26 | -1.2263582 | -1.1041391 | -1.1652487 | 0.1222191 | 2 | 9.249914 | 13.727711 | 0.0017043 | 0.0246056 | Endoplasmic reticulum resident protein 44 |
| Q9UKR5 | -3.2086830 | -4.2783690 | -3.7435260 | -1.0696861 | 2 | 8.595465 | 14.600416 | 0.0017210 | 0.0246056 | Probable ergosterol biosynthetic protein 28 |
| Q9HAV4 | -2.2052737 | -1.1596644 | -1.6824690 | 1.0456093 | 2 | 7.987893 | 15.661958 | 0.0017212 | 0.0246056 | Exportin-5 |
| Q9BTV4 | -1.8104632 | -1.4010178 | -1.6057405 | 0.4094454 | 2 | 9.284468 | 13.566939 | 0.0017561 | 0.0248698 | Transmembrane protein 43 |
| P35052 | -0.6193263 | -1.9557489 | -1.2875376 | -1.3364227 | 2 | 7.703314 | 16.101781 | 0.0017721 | 0.0248698 | Glypican-1;Secreted glypican-1 |
| P01008 | -1.5471922 | -1.5321558 | -1.5396740 | 0.0150364 | 2 | 9.310907 | 13.489881 | 0.0017764 | 0.0248698 | Antithrombin-III |
| P13671 | -1.1914382 | -1.5383124 | -1.3648753 | -0.3468742 | 2 | 9.385413 | 13.329057 | 0.0018101 | 0.0249035 | Complement component C6 |
| P14543 | -1.3845506 | -1.2999894 | -1.3422700 | 0.0845612 | 2 | 9.204669 | 13.536114 | 0.0018145 | 0.0249035 | Nidogen-1 |
| Q12996 | -1.6674028 | -1.2935582 | -1.4804805 | 0.3738446 | 2 | 7.207709 | 17.128176 | 0.0018261 | 0.0249035 | Cleavage stimulation factor subunit 3 |
| Q2TAA5 | -1.3192452 | -0.9694554 | -1.1443503 | 0.3497898 | 2 | 9.249693 | 13.452203 | 0.0018279 | 0.0249035 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase |
| Q6PI78 | 1.1229874 | 1.9484794 | 1.5357334 | 0.8254920 | 2 | 9.040261 | 13.666187 | 0.0018499 | 0.0249839 | Transmembrane protein 65 |
| P01042 | -2.2563158 | -1.7570236 | -2.0066697 | 0.4992922 | 2 | 7.210294 | 17.020705 | 0.0018584 | 0.0249839 | Kininogen-1;Kininogen-1 heavy chain;T-kinin;Bradykinin;Lysyl-bradykinin;Kininogen-1 light chain;Low molecular weight growth-promoting factor |
| Q96H79 | -3.0216701 | -1.3331324 | -2.1774013 | 1.6885377 | 2 | 6.334231 | 19.561690 | 0.0019464 | 0.0259943 | Zinc finger CCCH-type antiviral protein 1-like |
| Q8TDB4 | -1.8386665 | -2.8602879 | -2.3494772 | -1.0216214 | 2 | 6.330306 | 19.494450 | 0.0019690 | 0.0261246 | Protein MGARP |
| P45877 | -1.2597920 | -0.7394688 | -0.9996304 | 0.5203232 | 2 | 9.343571 | 13.002295 | 0.0019971 | 0.0261892 | Peptidyl-prolyl cis-trans isomerase C |
| P20774 | -2.2367735 | -1.9653377 | -2.1010556 | 0.2714358 | 2 | 8.210882 | 14.549345 | 0.0019997 | 0.0261892 | Mimecan |
| Q9HAT2 | 1.3442519 | 1.8668820 | 1.6055669 | 0.5226301 | 2 | 9.216677 | 13.044682 | 0.0020515 | 0.0266959 | Sialate O-acetylesterase |
| Q9UJC5 | -1.1262301 | -1.3257262 | -1.2259781 | -0.1994961 | 2 | 9.326832 | 12.870624 | 0.0020783 | 0.0268689 | SH3 domain-binding glutamic acid-rich-like protein 2 |
| P34932 | -1.1634292 | -0.6217632 | -0.8925962 | 0.5416660 | 2 | 9.385413 | 12.783000 | 0.0020913 | 0.0268689 | Heat shock 70 kDa protein 4 |
| Q9GZY4 | -3.1677681 | -0.0954235 | -1.6315958 | 3.0723446 | 2 | 7.703311 | 15.142943 | 0.0021422 | 0.0271006 | Cytochrome c oxidase assembly factor 1 homolog |
| Q96C86 | -1.5155718 | -0.2524683 | -0.8840200 | 1.2631035 | 2 | 9.145534 | 12.949577 | 0.0021488 | 0.0271006 | m7GpppX diphosphatase |
| Q16082 | -1.3619821 | -1.2847033 | -1.3233427 | 0.0772788 | 2 | 9.207589 | 12.877393 | 0.0021494 | 0.0271006 | Heat shock protein beta-2 |
| Q96KC8 | 0.8980074 | -2.5287840 | -0.8153883 | -3.4267914 | 2 | 8.321700 | 13.939197 | 0.0022045 | 0.0276241 | DnaJ homolog subfamily C member 1 |
| P56539 | -2.0519177 | 0.6926641 | -0.6796268 | 2.7445818 | 2 | 8.553679 | 13.356405 | 0.0023380 | 0.0291177 | Caveolin-3 |
| P62857 | -0.6453035 | -2.1384365 | -1.3918700 | -1.4931330 | 2 | 7.956911 | 14.197256 | 0.0023720 | 0.0293602 | 40S ribosomal protein S28 |
| P06727 | -1.3583680 | -0.7667083 | -1.0625381 | 0.5916597 | 2 | 8.884167 | 12.814824 | 0.0024096 | 0.0295719 | Apolipoprotein A-IV |
| P02461 | -3.5976656 | -2.2738894 | -2.9357775 | 1.3237762 | 2 | 8.323445 | 13.522376 | 0.0024274 | 0.0295719 | Collagen alpha-1(III) chain |
| Q14314 | -1.7692180 | -2.0953097 | -1.9322639 | -0.3260917 | 2 | 9.382448 | 12.231706 | 0.0024328 | 0.0295719 | Fibroleukin |
| P09619 | -1.6757284 | -0.9093138 | -1.2925211 | 0.7664146 | 2 | 9.154490 | 12.419073 | 0.0024667 | 0.0298056 | Platelet-derived growth factor receptor beta |
| O15061 | -0.4856308 | -1.7028211 | -1.0942259 | -1.2171903 | 2 | 9.385413 | 12.086427 | 0.0025311 | 0.0304031 | Synemin |
| P19429 | 2.0373090 | 2.1009541 | 2.0691315 | 0.0636451 | 2 | 7.976624 | 13.699861 | 0.0026302 | 0.0313664 | Troponin I, cardiac muscle |
| P02790 | -1.1921329 | -1.4568633 | -1.3244981 | -0.2647304 | 2 | 9.385413 | 11.933537 | 0.0026422 | 0.0313664 | Hemopexin |
| P01034 | -1.6893296 | -1.2428420 | -1.4660858 | 0.4464875 | 2 | 8.837162 | 12.445434 | 0.0026903 | 0.0317523 | Cystatin-C |
| P01699 | -4.0259605 | 0.3400879 | -1.8429363 | 4.3660484 | 2 | 6.314916 | 17.240217 | 0.0027650 | 0.0323664 | Ig lambda chain V-I region VOR |
| Q07507 | -0.9566123 | -1.6741554 | -1.3153838 | -0.7175431 | 2 | 8.160106 | 13.191734 | 0.0027743 | 0.0323664 | Dermatopontin |
| P46063 | -1.3869124 | -0.2234828 | -0.8051976 | 1.1634296 | 2 | 9.154885 | 11.934461 | 0.0028156 | 0.0326607 | ATP-dependent DNA helicase Q1 |
| Q9Y2Z0 | -1.7579777 | 1.2068202 | -0.2755788 | 2.9647978 | 2 | 9.385413 | 11.674630 | 0.0028441 | 0.0327268 | Suppressor of G2 allele of SKP1 homolog |
| P50479 | -1.9052893 | -2.3347923 | -2.1200408 | -0.4295030 | 2 | 8.131897 | 13.113069 | 0.0028535 | 0.0327268 | PDZ and LIM domain protein 4 |
| Q86VP6 | -1.0974174 | -1.6122394 | -1.3548284 | -0.5148220 | 2 | 8.542953 | 12.540888 | 0.0028725 | 0.0327590 | Cullin-associated NEDD8-dissociated protein 1 |
| P28300 | -1.2607685 | -1.6978873 | -1.4793279 | -0.4371188 | 2 | 7.649016 | 13.768131 | 0.0029194 | 0.0330790 | Protein-lysine 6-oxidase |
| Q15126 | -1.0340183 | -0.9865904 | -1.0103044 | 0.0474279 | 2 | 9.342811 | 11.607013 | 0.0029331 | 0.0330790 | Phosphomevalonate kinase |
| Q86VU5 | 1.5740900 | 1.1899321 | 1.3820110 | -0.3841579 | 2 | 9.385413 | 11.512614 | 0.0029801 | 0.0334227 | Catechol O-methyltransferase domain-containing protein 1 |
| O43143 | -1.0109780 | -0.7834921 | -0.8972350 | 0.2274859 | 2 | 9.289616 | 11.574983 | 0.0030027 | 0.0334915 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 |
| P82663 | -0.7938408 | -1.9350886 | -1.3644647 | -1.1412478 | 2 | 9.302714 | 11.525853 | 0.0030347 | 0.0336069 | 28S ribosomal protein S25, mitochondrial |
| O43175 | -2.0890071 | -2.0660744 | -2.0775407 | 0.0229327 | 2 | 9.173117 | 11.629663 | 0.0030510 | 0.0336069 | D-3-phosphoglycerate dehydrogenase |
| P26447 | -1.8777689 | -1.1686882 | -1.5232285 | 0.7090807 | 2 | 8.774968 | 12.025412 | 0.0030627 | 0.0336069 | Protein S100-A4 |
| P13667 | -1.1592112 | -1.0499894 | -1.1046003 | 0.1092218 | 2 | 9.181460 | 11.588346 | 0.0030799 | 0.0336143 | Protein disulfide-isomerase A4 |
| Q04760 | 0.5615089 | 2.2161884 | 1.3888487 | 1.6546795 | 2 | 8.599181 | 12.147102 | 0.0031249 | 0.0338146 | Lactoylglutathione lyase |
| Q96CS3 | -1.3291851 | -1.0516357 | -1.1904104 | 0.2775494 | 2 | 9.119924 | 11.588823 | 0.0031316 | 0.0338146 | FAS-associated factor 2 |
| P50552 | -1.5231187 | -0.3481323 | -0.9356255 | 1.1749865 | 2 | 9.074476 | 11.556793 | 0.0031996 | 0.0343659 | Vasodilator-stimulated phosphoprotein |
| O95183 | -1.3898689 | -1.5291947 | -1.4595318 | -0.1393258 | 2 | 9.371170 | 11.184431 | 0.0032924 | 0.0351768 | Vesicle-associated membrane protein 5 |
| Q15582 | -2.0883637 | -2.4627789 | -2.2755713 | -0.3744151 | 2 | 7.899813 | 12.560835 | 0.0035188 | 0.0370726 | Transforming growth factor-beta-induced protein ig-h3 |
| Q6UWS5 | 0.9342388 | 1.7762603 | 1.3552496 | 0.8420216 | 2 | 7.068224 | 13.945225 | 0.0035193 | 0.0370726 | Protein PET117 homolog, mitochondrial |
| P46940 | -1.2205560 | -1.0146605 | -1.1176083 | 0.2058955 | 2 | 9.306592 | 11.010224 | 0.0035246 | 0.0370726 | Ras GTPase-activating-like protein IQGAP1 |
| Q96JB2 | -1.5453539 | -2.4514355 | -1.9983947 | -0.9060817 | 2 | 8.142504 | 12.081209 | 0.0036586 | 0.0381425 | Conserved oligomeric Golgi complex subunit 3 |
| Q9H1E5 | -1.3888294 | -0.6312590 | -1.0100442 | 0.7575704 | 2 | 9.385413 | 10.814657 | 0.0036639 | 0.0381425 | Thioredoxin-related transmembrane protein 4 |
| P62328 | -1.2899569 | -1.6678529 | -1.4789049 | -0.3778960 | 2 | 7.220282 | 13.330964 | 0.0037678 | 0.0388339 | Thymosin beta-4;Hematopoietic system regulatory peptide |
| Q08945 | -2.0713524 | -0.7768868 | -1.4241196 | 1.2944656 | 2 | 8.385413 | 11.678065 | 0.0037686 | 0.0388339 | FACT complex subunit SSRP1 |
| Q5VIR6-4 | -1.7550575 | -0.0614140 | -0.9082357 | 1.6936435 | 2 | 9.130015 | 10.919027 | 0.0037890 | 0.0388469 | Vacuolar protein sorting-associated protein 53 homolog |
| P15924 | 1.2742208 | 0.5869741 | 0.9305974 | -0.6872468 | 2 | 9.350459 | 10.675543 | 0.0038552 | 0.0391584 | Desmoplakin |
| P01019 | -1.0362862 | -1.0127277 | -1.0245069 | 0.0235585 | 2 | 9.093510 | 10.889809 | 0.0038580 | 0.0391584 | Angiotensinogen;Angiotensin-1;Angiotensin-2;Angiotensin-3;Angiotensin-4;Angiotensin 1-9;Angiotensin 1-7;Angiotensin 1-5;Angiotensin 1-4 |
| P83916 | -1.9742521 | -1.3616457 | -1.6679489 | 0.6126064 | 2 | 6.385413 | 14.963203 | 0.0038901 | 0.0392877 | Chromobox protein homolog 1 |
| O95486 | -1.5003000 | -1.1213028 | -1.3108014 | 0.3789972 | 2 | 9.187800 | 10.736201 | 0.0039420 | 0.0396155 | Protein transport protein Sec24A |
| P03950 | -2.2142952 | -1.3775033 | -1.7958993 | 0.8367919 | 2 | 8.216113 | 11.647751 | 0.0039967 | 0.0399668 | Angiogenin |
| Q9NRX4 | 1.2804301 | 1.4870941 | 1.3837621 | 0.2066640 | 2 | 9.339127 | 10.535987 | 0.0040345 | 0.0400138 | 14 kDa phosphohistidine phosphatase |
| Q9Y5U8 | -3.9888327 | -0.8866380 | -2.4377353 | 3.1021947 | 2 | 9.327097 | 10.540498 | 0.0040408 | 0.0400138 | Mitochondrial pyruvate carrier 1 |
| Q8N5M1 | -2.0221430 | -0.6423210 | -1.3322320 | 1.3798221 | 2 | 8.937206 | 10.851063 | 0.0040640 | 0.0400479 | ATP synthase mitochondrial F1 complex assembly factor 2 |
| Q562R1 | 1.3201209 | -1.9616415 | -0.3207603 | -3.2817624 | 2 | 9.266665 | 10.545352 | 0.0040950 | 0.0401588 | Beta-actin-like protein 2 |
| P05455 | -1.2042291 | -0.5305125 | -0.8673708 | 0.6737166 | 2 | 9.178555 | 10.565065 | 0.0041602 | 0.0403946 | Lupus La protein |
| O15118 | -1.6169068 | -2.6334712 | -2.1251890 | -1.0165644 | 2 | 6.385413 | 14.553122 | 0.0041844 | 0.0403946 | Niemann-Pick C1 protein |
| Q04721 | 1.5310391 | 0.6944075 | 1.1127233 | -0.8366316 | 2 | 9.201253 | 10.522611 | 0.0041905 | 0.0403946 | Neurogenic locus notch homolog protein 2;Notch 2 extracellular truncation;Notch 2 intracellular domain |
| P54577 | -1.3097944 | -0.7452704 | -1.0275324 | 0.5645241 | 2 | 8.964139 | 10.716040 | 0.0041986 | 0.0403946 | Tyrosine–tRNA ligase, cytoplasmic;Tyrosine–tRNA ligase, cytoplasmic, N-terminally processed |
| P00747 | -0.7054831 | -0.9638979 | -0.8346905 | -0.2584148 | 2 | 9.172800 | 10.440542 | 0.0043269 | 0.0413862 | Plasminogen;Plasmin heavy chain A;Activation peptide;Angiostatin;Plasmin heavy chain A, short form;Plasmin light chain B |
| P02748 | -1.3394634 | -1.1229428 | -1.2312031 | 0.2165205 | 2 | 8.865762 | 10.685485 | 0.0043459 | 0.0413862 | Complement component C9;Complement component C9a;Complement component C9b |
| Q9BXR6 | -2.3526616 | -0.3476437 | -1.3501527 | 2.0050179 | 2 | 9.382448 | 10.250529 | 0.0043629 | 0.0413862 | Complement factor H-related protein 5 |
| Q7Z3T8 | -1.5768427 | -1.2158882 | -1.3963655 | 0.3609544 | 2 | 8.413206 | 11.050965 | 0.0044277 | 0.0417033 | Zinc finger FYVE domain-containing protein 16 |
| Q9Y287 | -1.9187015 | -1.3340175 | -1.6263595 | 0.5846840 | 2 | 9.385413 | 10.194354 | 0.0044376 | 0.0417033 | Integral membrane protein 2B;BRI2, membrane form;BRI2 intracellular domain;BRI2C, soluble form;Bri23 peptide |
| P01303 | -0.6538416 | -2.1633002 | -1.4085709 | -1.5094586 | 2 | 8.890312 | 10.577519 | 0.0044579 | 0.0417033 | Pro-neuropeptide Y;Neuropeptide Y;C-flanking peptide of NPY |
| Q8WWQ0 | -1.3655434 | -1.2188019 | -1.2921726 | 0.1467415 | 2 | 9.385413 | 10.140256 | 0.0045140 | 0.0420343 | PH-interacting protein |
| O95445 | -2.7585923 | -2.3195737 | -2.5390830 | 0.4390186 | 2 | 9.052359 | 10.351660 | 0.0045797 | 0.0421706 | Apolipoprotein M |
| P27658 | -1.5409469 | -1.7449610 | -1.6429540 | -0.2040141 | 2 | 9.385413 | 10.090334 | 0.0045860 | 0.0421706 | Collagen alpha-1(VIII) chain;Vastatin |
| Q9UK22 | -1.9689505 | -0.9835733 | -1.4762619 | 0.9853772 | 2 | 6.372396 | 14.069977 | 0.0045960 | 0.0421706 | F-box only protein 2 |
| P80723 | -1.6385395 | -1.4066562 | -1.5225978 | 0.2318833 | 2 | 7.910668 | 11.448438 | 0.0046195 | 0.0421706 | Brain acid soluble protein 1 |
| P01611 | -0.4068850 | -2.2194112 | -1.3131481 | -1.8125261 | 2 | 9.385413 | 10.058567 | 0.0046325 | 0.0421706 | Ig kappa chain V-I region Wes |
| Q9UBV8 | -1.1877604 | -1.1566251 | -1.1721927 | 0.0311353 | 2 | 8.885488 | 10.413473 | 0.0046866 | 0.0424726 | Peflin |
| Q9UKX3 | 2.0400959 | 1.3472678 | 1.6936818 | -0.6928281 | 2 | 9.208912 | 10.120583 | 0.0047349 | 0.0427193 | Myosin-13 |
| Q8TDB6 | -1.6113146 | -0.2207985 | -0.9160565 | 1.3905161 | 2 | 9.153547 | 10.138005 | 0.0047718 | 0.0428622 | E3 ubiquitin-protein ligase DTX3L |
| P07360 | -0.3859901 | -1.5479619 | -0.9669760 | -1.1619718 | 2 | 8.752393 | 10.443655 | 0.0048053 | 0.0429727 | Complement component C8 gamma chain |
| P08582 | -1.6638598 | 0.4414480 | -0.6112059 | 2.1053079 | 2 | 9.299314 | 9.966955 | 0.0048656 | 0.0432000 | Melanotransferrin |
| O60262 | -1.5147797 | -0.9563725 | -1.2355761 | 0.5584072 | 2 | 8.388576 | 10.731124 | 0.0048733 | 0.0432000 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-7 |
| P52907 | -0.6186589 | -1.0777393 | -0.8481991 | -0.4590804 | 2 | 9.339311 | 9.904666 | 0.0049178 | 0.0432527 | F-actin-capping protein subunit alpha-1 |
| P07384 | -1.1069382 | -0.6510097 | -0.8789740 | 0.4559285 | 2 | 9.333139 | 9.906524 | 0.0049219 | 0.0432527 | Calpain-1 catalytic subunit |
| P00352 | -1.1650439 | -1.1023149 | -1.1336794 | 0.0627289 | 2 | 8.402816 | 10.653653 | 0.0049615 | 0.0434132 | Retinal dehydrogenase 1 |
| Q6P1N0 | -1.6996040 | -1.6333624 | -1.6664832 | 0.0662416 | 2 | 6.936107 | 12.521980 | 0.0049892 | 0.0434683 | Coiled-coil and C2 domain-containing protein 1A |
| O75348 | -1.8464170 | -0.7442245 | -1.2953207 | 1.1021925 | 2 | 9.377764 | 9.811092 | 0.0050240 | 0.0434758 | V-type proton ATPase subunit G 1 |
| P14555 | -4.5949034 | -1.7126693 | -3.1537863 | 2.8822341 | 2 | 6.867951 | 12.600143 | 0.0050329 | 0.0434758 | Phospholipase A2, membrane associated |
| Q9UKS6 | 1.4436881 | 0.5239624 | 0.9838252 | -0.9197256 | 2 | 8.943424 | 10.075794 | 0.0051177 | 0.0438921 | Protein kinase C and casein kinase substrate in neurons protein 3 |
| Q9BSD7 | -0.1234697 | 2.9508964 | 1.4137133 | 3.0743661 | 2 | 7.080846 | 12.162214 | 0.0051243 | 0.0438921 | Cancer-related nucleoside-triphosphatase |
| Q9BYN0 | -1.2973636 | -1.2743737 | -1.2858687 | 0.0229898 | 2 | 9.180640 | 9.864374 | 0.0051672 | 0.0440735 | Sulfiredoxin-1 |
| P04350 | -0.5004642 | -2.8380383 | -1.6692512 | -2.3375741 | 2 | 6.967833 | 12.257034 | 0.0052259 | 0.0443875 | Tubulin beta-4A chain |
| Q9BXV9 | 1.5862723 | 1.2391702 | 1.4127212 | -0.3471021 | 2 | 9.163202 | 9.797960 | 0.0052989 | 0.0448202 | Uncharacterized protein C14orf142 |
| Q9UL26 | -1.2168882 | 7.9238349 | 3.3534734 | 9.1407231 | 2 | 8.784585 | 10.041159 | 0.0053777 | 0.0451937 | Ras-related protein Rab-22A |
| Q96A65 | -0.6560056 | -1.0764957 | -0.8662506 | -0.4204901 | 2 | 9.382448 | 9.593458 | 0.0053876 | 0.0451937 | Exocyst complex component 4 |
| P24311 | 1.7257653 | 0.4145062 | 1.0701358 | -1.3112591 | 2 | 9.295713 | 9.617255 | 0.0054499 | 0.0454918 | Cytochrome c oxidase subunit 7B, mitochondrial |
| P62745 | -1.4954700 | -0.9675811 | -1.2315256 | 0.5278889 | 2 | 8.788016 | 9.983785 | 0.0054680 | 0.0454918 | Rho-related GTP-binding protein RhoB |
| A5D6W6 | -2.3613596 | -2.0136134 | -2.1874865 | 0.3477462 | 2 | 7.868778 | 10.815729 | 0.0055206 | 0.0457417 | Fat storage-inducing transmembrane protein 1 |
| Q8IYI6 | -1.0552142 | -0.8759545 | -0.9655844 | 0.1792596 | 2 | 9.385413 | 9.452880 | 0.0056398 | 0.0465401 | Exocyst complex component 8 |
| Q8IYU8 | -2.9245934 | -3.1789343 | -3.0517638 | -0.2543409 | 2 | 6.385413 | 12.905411 | 0.0057115 | 0.0468485 | Calcium uptake protein 2, mitochondrial |
| Q6IC98 | -1.1266523 | -0.6748287 | -0.9007405 | 0.4518236 | 2 | 8.888308 | 9.747386 | 0.0057422 | 0.0468485 | GRAM domain-containing protein 4 |
| Q8N142 | 0.9014180 | 1.2636791 | 1.0825486 | 0.3622611 | 2 | 9.201572 | 9.519259 | 0.0057464 | 0.0468485 | Adenylosuccinate synthetase isozyme 1 |
| P50991 | -1.7861491 | -1.1833510 | -1.4847501 | 0.6027981 | 2 | 8.685140 | 9.847405 | 0.0058465 | 0.0474077 | T-complex protein 1 subunit delta |
| Q01581 | -0.9331228 | -2.3288187 | -1.6309708 | -1.3956959 | 2 | 7.186730 | 11.425555 | 0.0058617 | 0.0474077 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic |
| O00625 | -1.3732244 | -0.9202215 | -1.1467230 | 0.4530029 | 2 | 9.385413 | 9.307306 | 0.0059204 | 0.0476208 | Pirin |
| P25311 | -1.4767985 | -0.6753214 | -1.0760599 | 0.8014770 | 2 | 8.673237 | 9.807893 | 0.0059350 | 0.0476208 | Zinc-alpha-2-glycoprotein |
| O75746 | -1.5784821 | 0.3782752 | -0.6001034 | 1.9567573 | 2 | 8.893946 | 9.603894 | 0.0059996 | 0.0479394 | Calcium-binding mitochondrial carrier protein Aralar1 |
| P62277 | -1.5716452 | -1.0577672 | -1.3147062 | 0.5138780 | 2 | 9.269754 | 9.331127 | 0.0060219 | 0.0479394 | 40S ribosomal protein S13 |
| Q96GK7 | -1.0910279 | -1.4460031 | -1.2685155 | -0.3549752 | 2 | 7.917392 | 10.382537 | 0.0061228 | 0.0485522 | Fumarylacetoacetate hydrolase domain-containing protein 2A |
| Q14019 | -3.9736692 | -0.8629223 | -2.4182957 | 3.1107469 | 2 | 6.385413 | 12.535016 | 0.0061522 | 0.0485950 | Coactosin-like protein |
| P61009 | -1.0881392 | -0.6944647 | -0.8913019 | 0.3936745 | 2 | 8.621704 | 9.660391 | 0.0062888 | 0.0494818 | Signal peptidase complex subunit 3 |
| P36551 | -1.3385428 | 0.0293104 | -0.6546162 | 1.3678532 | 2 | 9.112223 | 9.290120 | 0.0063182 | 0.0495212 | Oxygen-dependent coproporphyrinogen-III oxidase, mitochondrial |
| Q96CX2 | -0.8346353 | -0.9677810 | -0.9012081 | -0.1331456 | 2 | 9.290787 | 9.153981 | 0.0063604 | 0.0496603 | BTB/POZ domain-containing protein KCTD12 |
| P19447 | -1.4553757 | -1.7864940 | -1.6209348 | -0.3311183 | 2 | 7.385413 | 10.812410 | 0.0063956 | 0.0496608 | TFIIH basal transcription factor complex helicase XPB subunit |
| Q9NS69 | -1.0286368 | -1.1141766 | -1.0714067 | -0.0855398 | 2 | 8.946979 | 9.359253 | 0.0064094 | 0.0496608 | Mitochondrial import receptor subunit TOM22 homolog |
5.3.3 Confirmation stage
##
## Attaching package: 'stageR'## The following object is masked from 'package:methods':
##
## getMethodpAll <- sapply(
c("omnibusTest",
"tissueV",
"tissueV + locationR:tissueV",
"tissueV + 0.5 * locationR:tissueV",
"locationR:tissueV"),
function(name,assay)
pull(rowData(assay)[,name],"pval"),
assay = pe[["proteinRobust"]])
rownames(pAll) <- rownames(pe[["proteinRobust"]])
stageWiseAnalysis <- stageR(pAll[,1], pAll[,-1])
stageWiseAnalysis <- stageWiseAdjustment(stageWiseAnalysis, method = "holm", alpha = 0.05, allowNA = TRUE)## Removing 1263 features with NA screening hypothesis p-values.rowData(pe[["proteinRobust"]])$stageWiseAnalysis <- data.frame(pAll)
rowData(pe[["proteinRobust"]])$stageWiseAnalysis[] <- NA
rowData(pe[["proteinRobust"]])$stageWiseAnalysis[getPScreen(stageWiseAnalysis) %>% is.na %>% `!`,] <- getAdjustedPValues(stageWiseAnalysis, onlySignificantGenes=FALSE, order=FALSE)## The returned adjusted p-values are based on a stage-wise testing approach and are only valid for the provided target OFDR level of 5%. If a different target OFDR level is of interest,the entire adjustment should be re-run.sigNamesStageWise <- apply(rowData(pe[["proteinRobust"]])$stageWiseAnalysis, 2, function(x) (x < 0.05) %>% which %>% names)
sapply(sigNamesStageWise,length)## omnibusTest tissueV
## 262 130
## tissueV...locationR.tissueV tissueV...0.5...locationR.tissueV
## 95 221
## locationR.tissueV
## 13for (i in 1:length(sigNamesStageWise))
heatmap(assay(pe[["proteinRobust"]])[sigNamesStageWise[[i]], ], main=names(sigNamesStageWise)[i])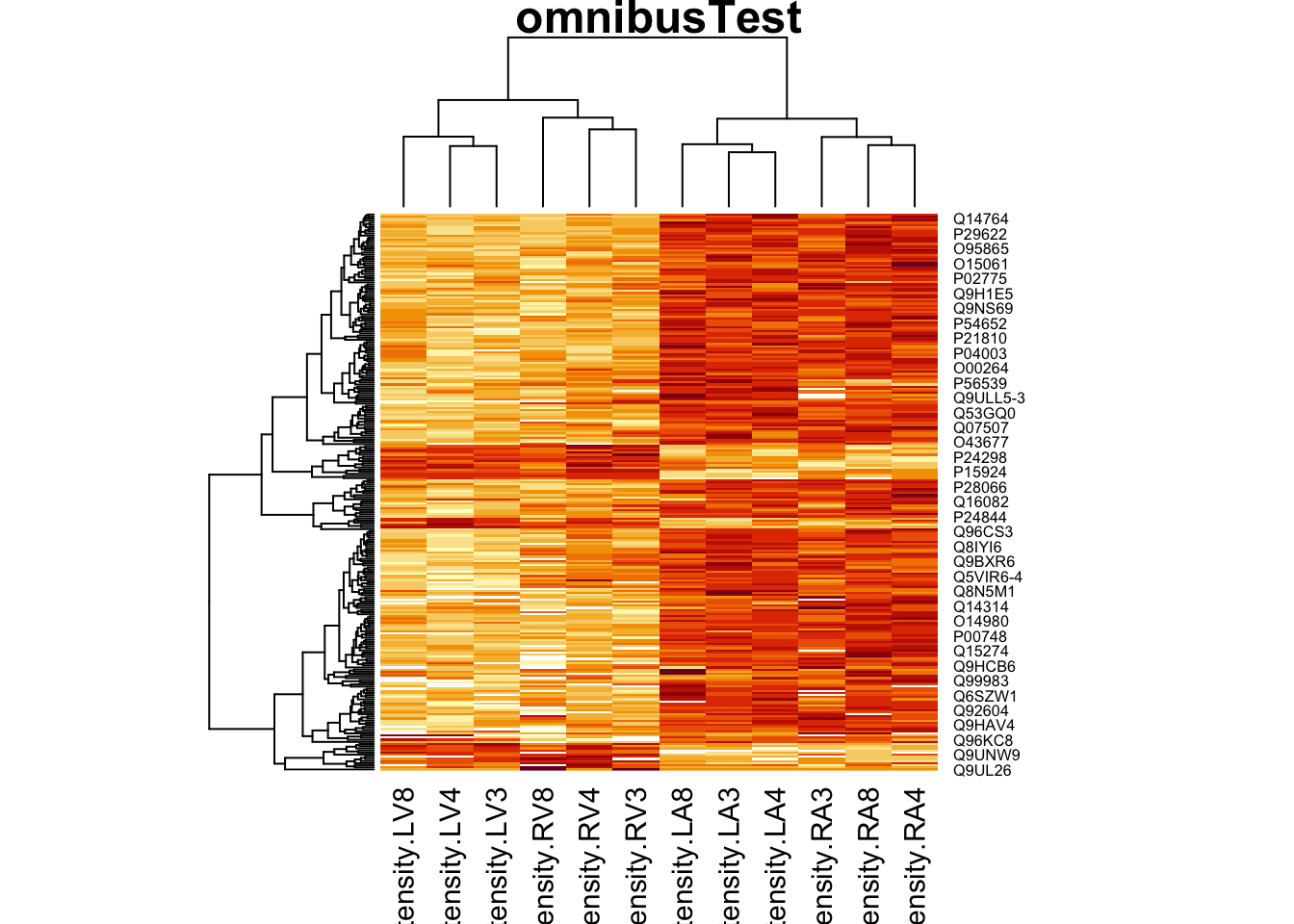
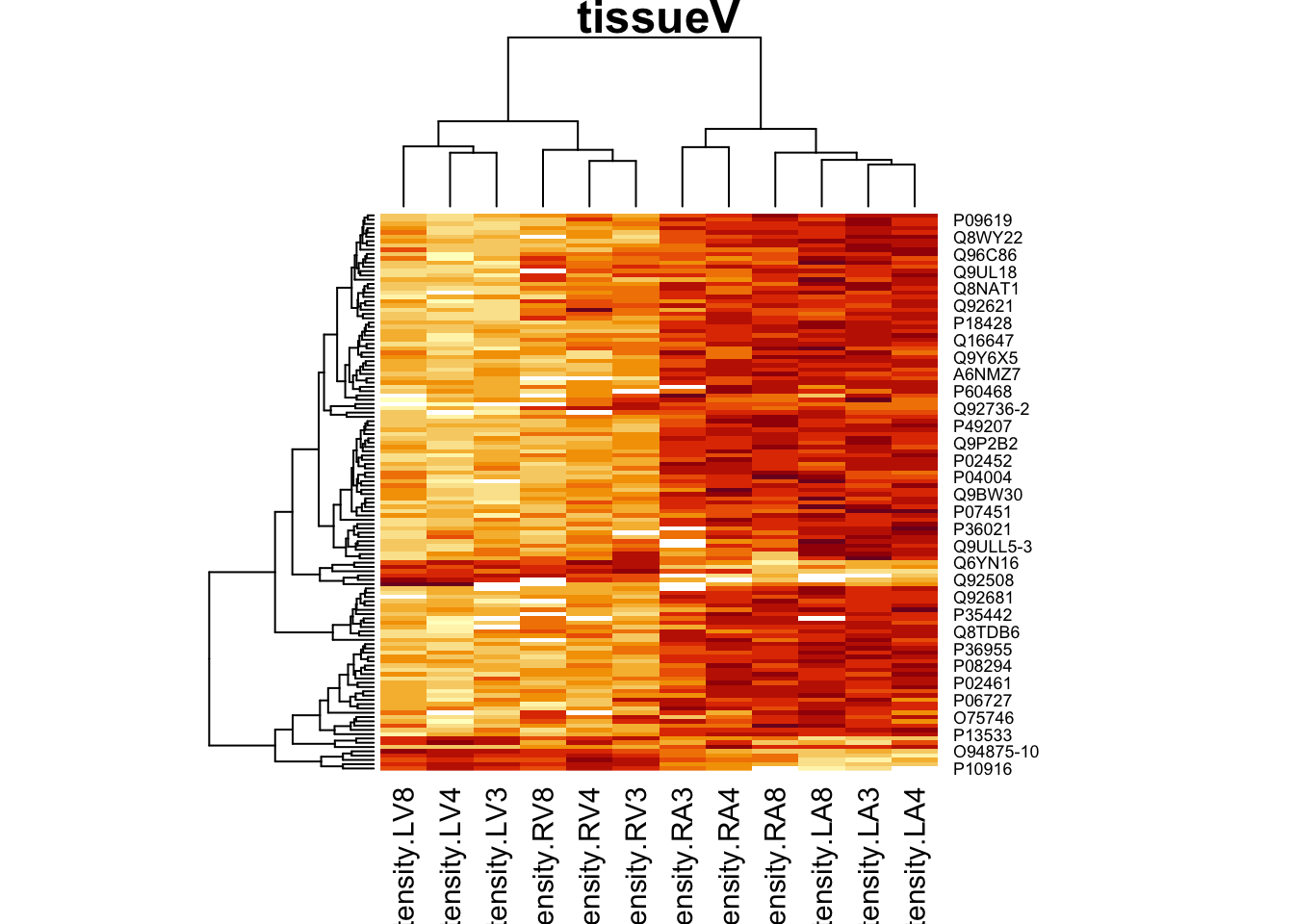
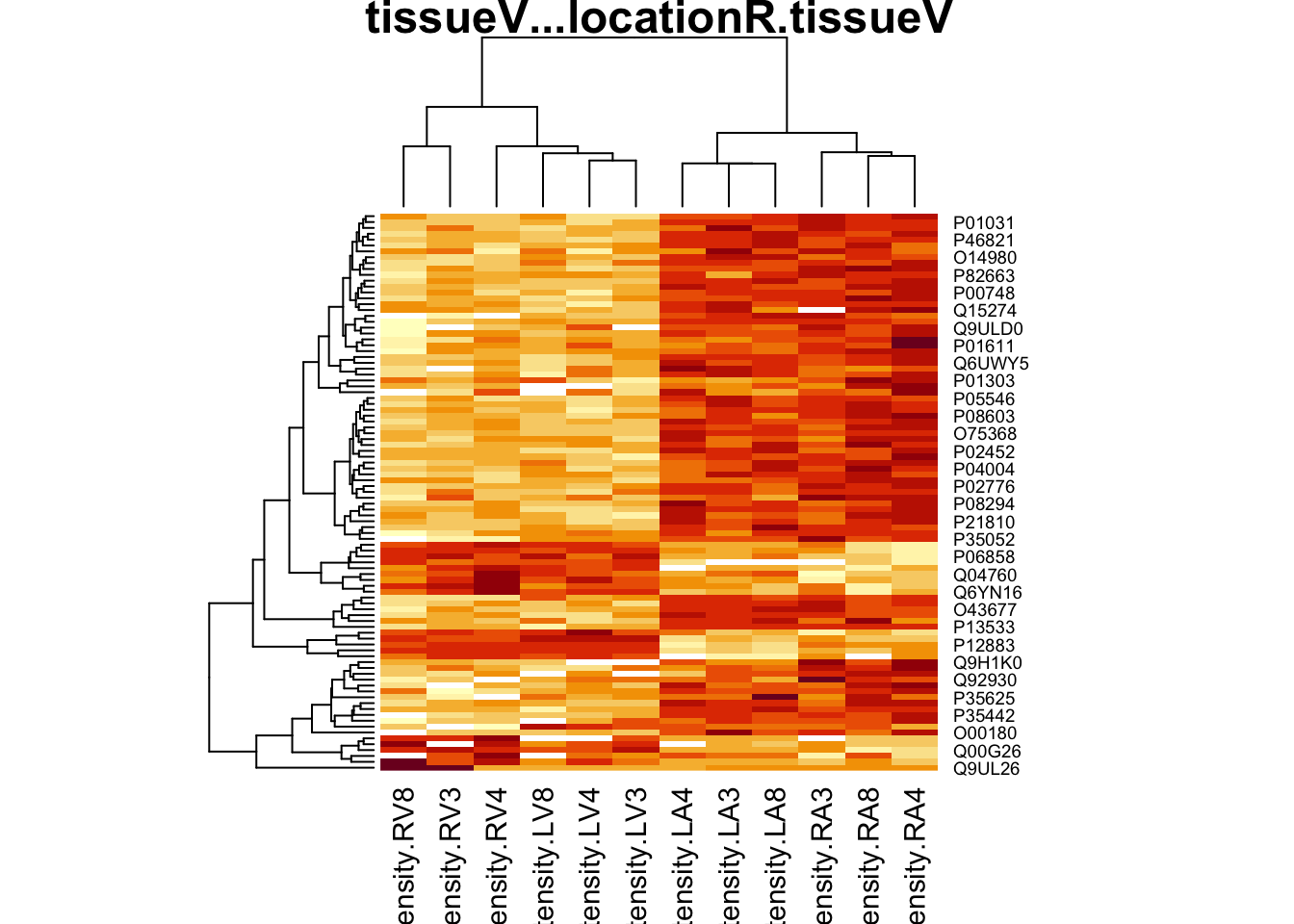
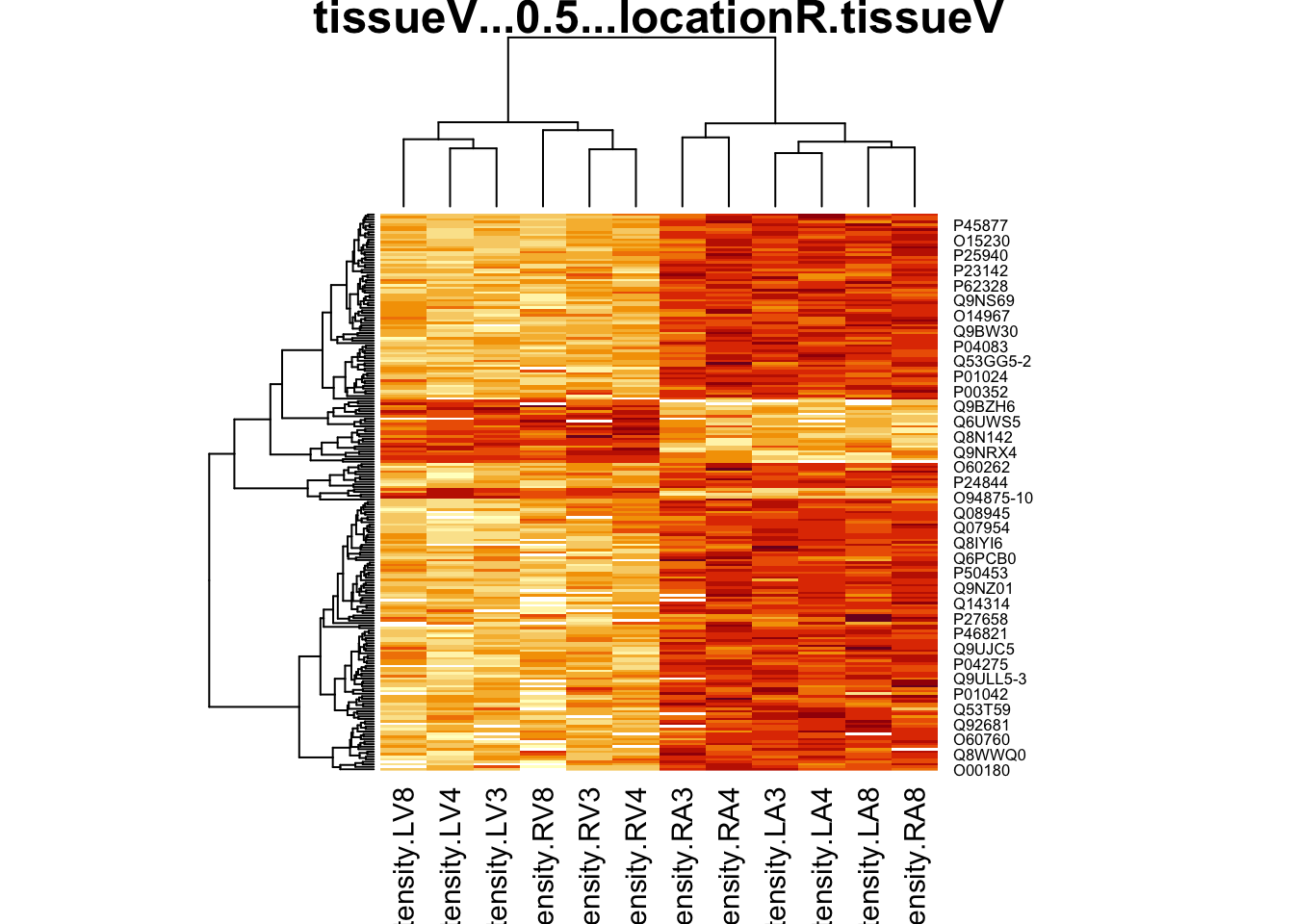
5.4 Confirmation improved
Instead of the Holm correction, we can use a better correction for this specific design in the confirmation stage. If we pass the screening stage we know that at least one null hypothesis related to the contrasts is false. If this is not the case we have made an false positive conclusion, but the multiple testing correction in the screening stage already accounts for that.
In the confirmation stage we have to correct for the maximum number of hypotheses that we can falsely reject. The omnibus hypothesis test essentially tests the overall null hypothesis that the log2 protein abundance is on average equal between atrium and ventriculum both in the left and right heart region. This involves testing simultaneously two model parameters: the main effect for ventriculum and the ventriculum:location interaction. All our hypotheses are based on these two model parameters, so the tests are not independent. The following configurations can occur: - None of the model parameters differ from zero, in which we can falsely reject all hypotheses. However, we already corrected for that in the screening stage! - If only the main ventriculum effect differs from zero, we know that the average log2 fold change between atrium and ventriculum left, right and overall differ from zero. So we falsely reject at most one null hypothesis, i.e. the interaction. - If only interaction differs from zero, only the null hypothesis related to log2 fold change in the left heart region can be falsely rejected. - If both the main effect and the interaction differ from zero it is possible that - the interaction is as large in magnitude as the main effect but opposite in sign. This would mean that the log2 fold change in the right heart region is zero, which we can falsely reject. - the interaction effect is twice as large in magnitude as the main effect but opposite in sign. This would mean that the log2 fold change in left and right side are equal in size but opposite in sign, implying that contrast that averages over the average log2 fold changes left and right would be zeor, which we could falsely reject - all effects are true.
Hence, as soon as we enter the confirmation stage that we can at maximum make one false rejection so for this design and for our hypotheses of interest we do not have to correct for multiple testing in the second stage because we only have to account for the fact that we can at most make one false rejection.
So we can set the method="none"
stageWiseAnalysis <- stageWiseAdjustment(stageWiseAnalysis, method = "none", alpha = 0.05, allowNA = TRUE)## Removing 1263 features with NA screening hypothesis p-values.rowData(pe[["proteinRobust"]])$stageWiseAnalysis <- data.frame(pAll)
rowData(pe[["proteinRobust"]])$stageWiseAnalysis[] <- NA
rowData(pe[["proteinRobust"]])$stageWiseAnalysis[getPScreen(stageWiseAnalysis) %>% is.na %>% `!`,] <- getAdjustedPValues(stageWiseAnalysis, onlySignificantGenes=FALSE, order=FALSE)## The returned adjusted p-values are based on a stage-wise testing approach and are only valid for the provided target OFDR level of 5%. If a different target OFDR level is of interest,the entire adjustment should be re-run.sigNamesStageWise <- apply(rowData(pe[["proteinRobust"]])$stageWiseAnalysis, 2, function(x) (x < 0.05) %>% which %>% names)
sapply(sigNamesStageWise,length)## omnibusTest tissueV
## 262 183
## tissueV...locationR.tissueV tissueV...0.5...locationR.tissueV
## 145 248
## locationR.tissueV
## 18for (i in 1:length(sigNamesStageWise))
heatmap(assay(pe[["proteinRobust"]])[sigNamesStageWise[[i]], ], main=names(sigNamesStageWise)[i])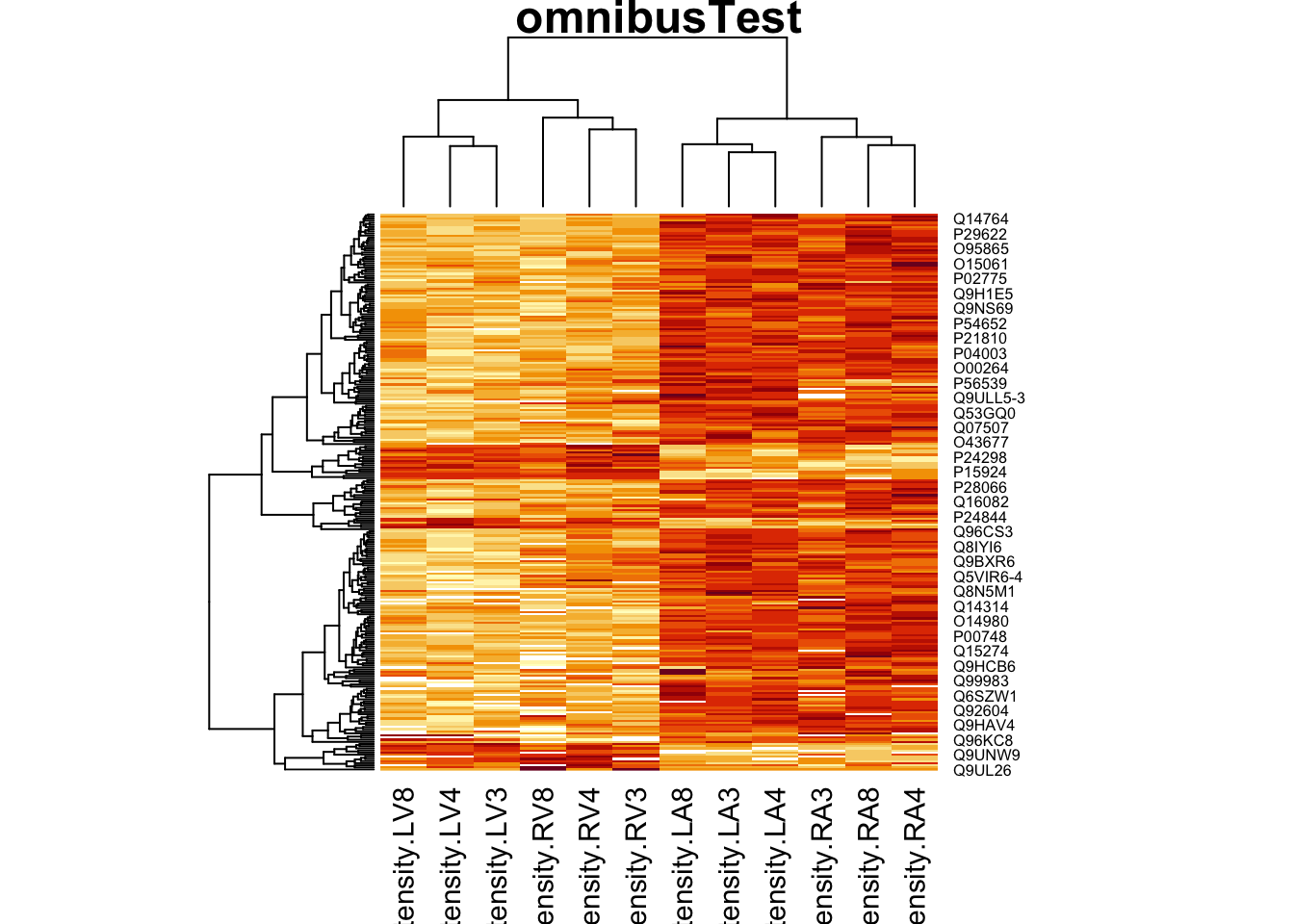
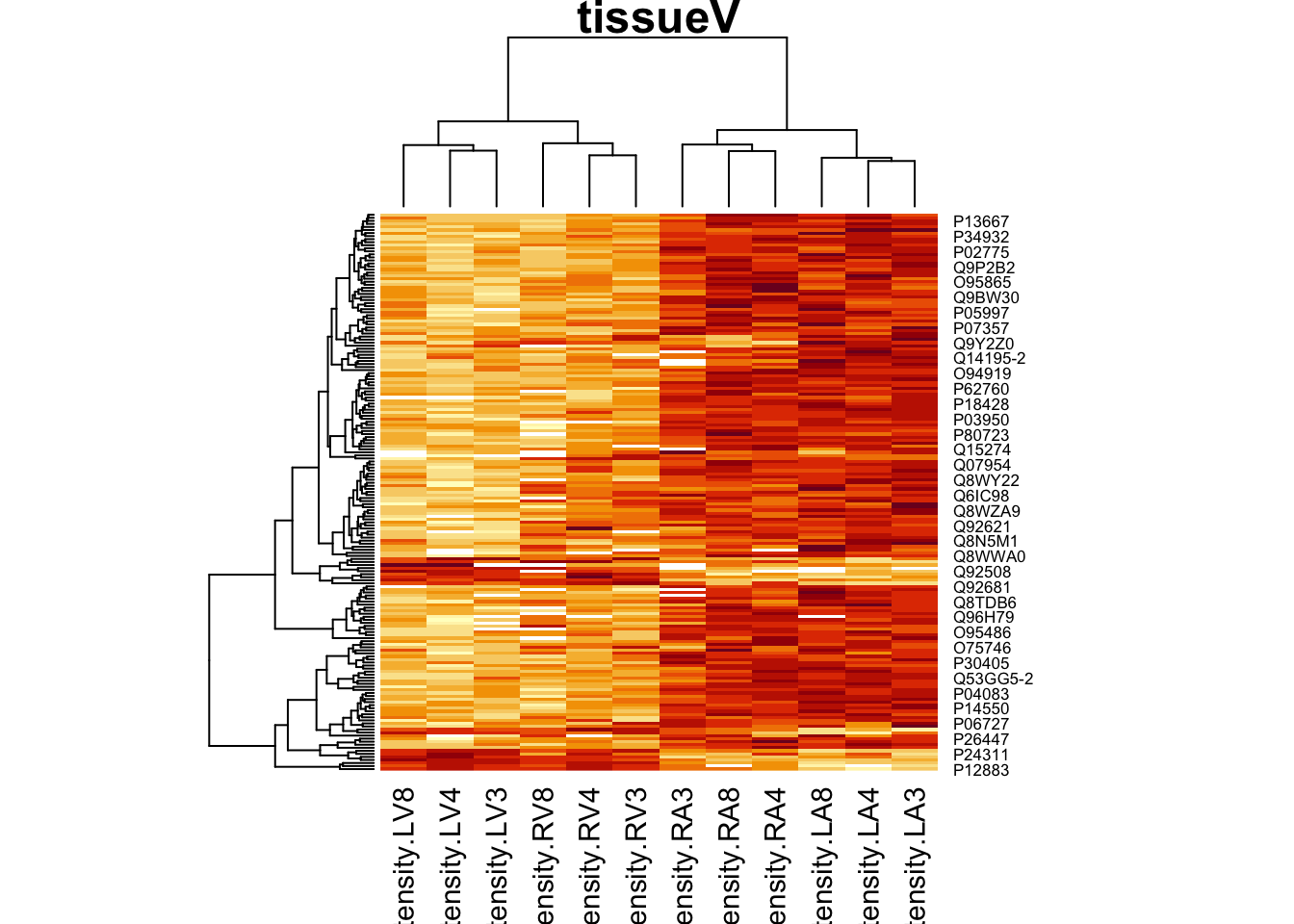
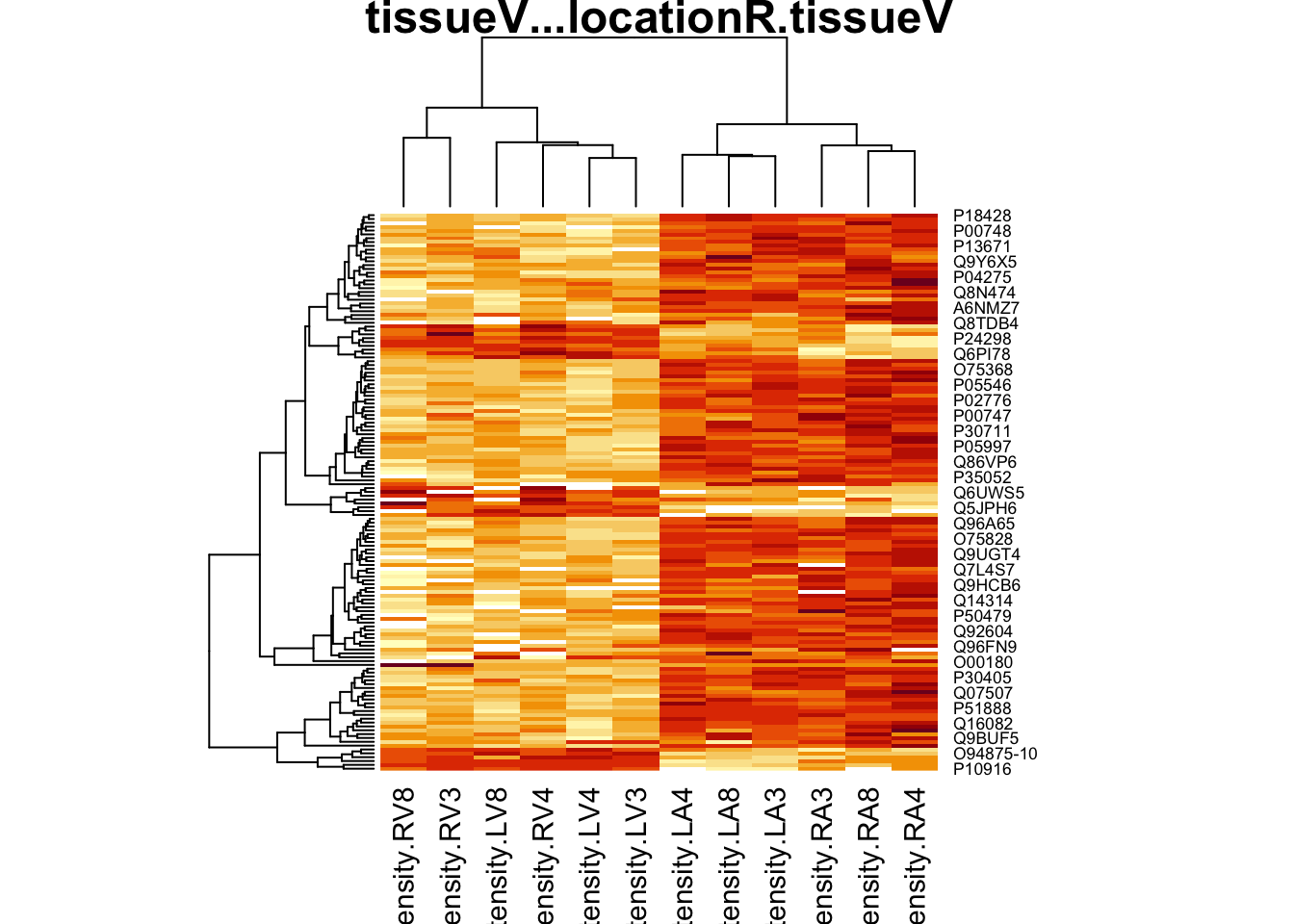
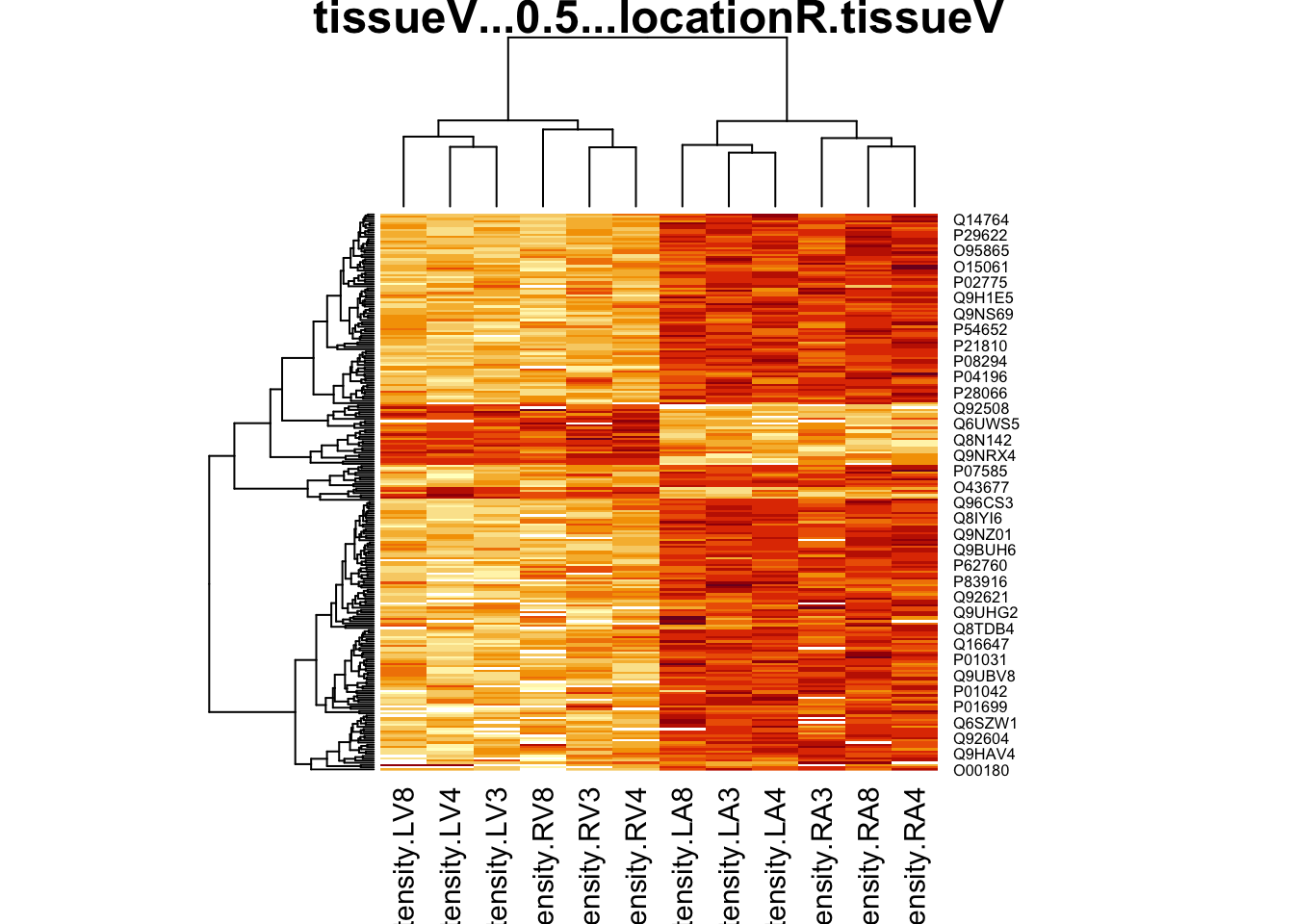
Note, that - our more relaxed FDR correction enables us to reject more null hypotheses in the second stage. - we can recover 18 proteins with significant interactions.
It is also very important to realise that setting the method to correct for multiple testing in the second stage at method = none is only correct for very specific designs and research hypotheses that are assessed. For most analyses we suggest to use the Holm method, which always we provide correct results.