Students are required to bring their own laptop with R version 3.6.0 or greater.
Please make sure that your computer’s hardware is sufficiently powered (>4 GB RAM, > 2 GB free disk space), that you have a working wireless card, and that you have administrator rights.
There are three options to work with the software:
- Install R/RStudio locally
- Using an online binder docker environment that launches R studio immediately. Remark: This tends to be unstable in combination with shiny Apps, the App gets disconnected when there is no browser activity in the App window.
1. Local installation
- Install R 3.6.0 or greater R/CRAN
- Install the latest version of Rstudio R/Rstudio
- To install all required packages, please copy and paste this line of code in your R console.
source("https://raw.githubusercontent.com/statOmics/SGA2020/master/install.R")
- Download and unzip SGA2019 master tree of the course to have the latest versions of the shiny apps.
- Go to the SGA2019 site on github: https://github.com/statOmics/SGA2019
- Click on the clone/download button and select download zip
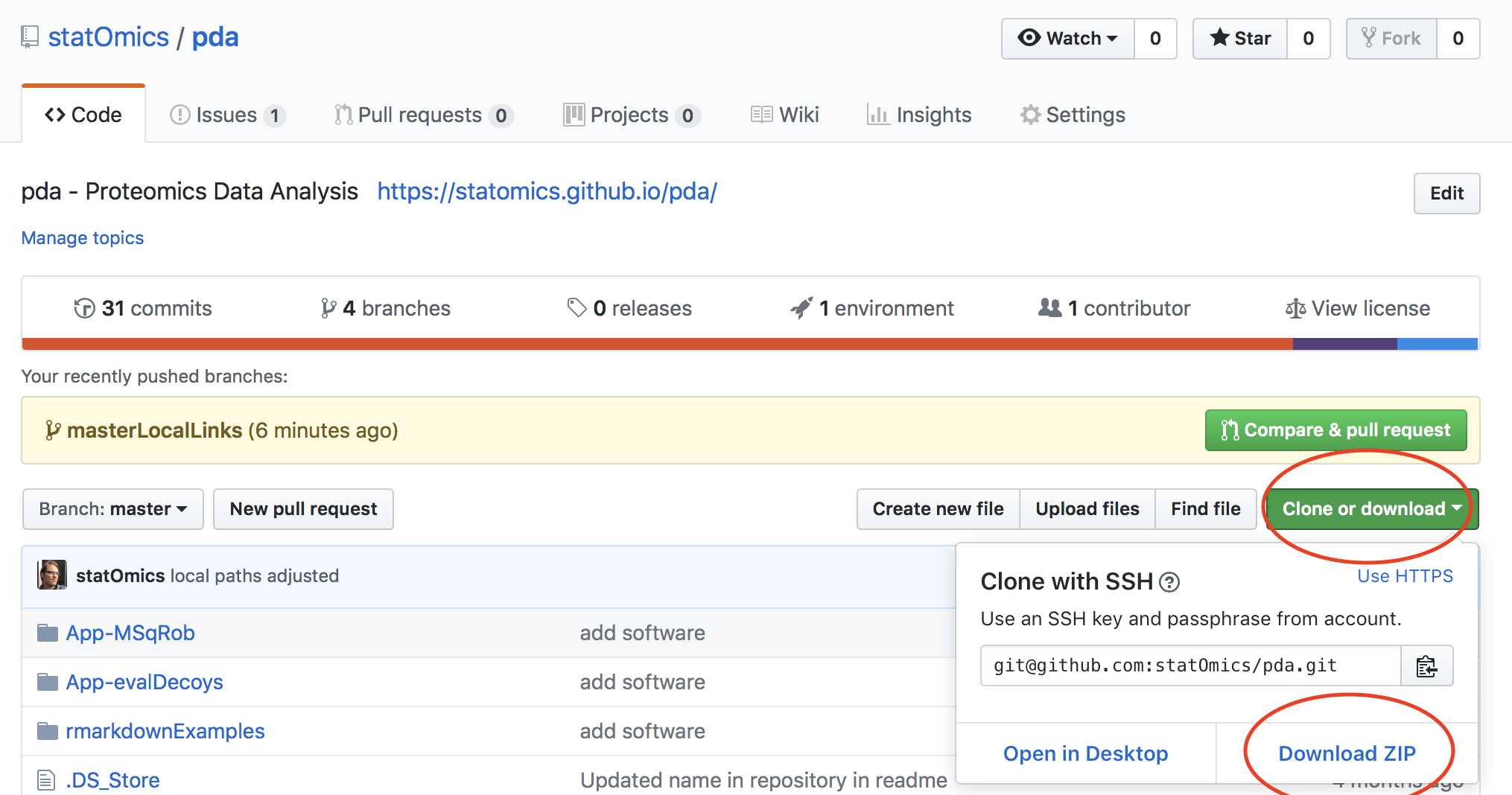
- Unzip the repository
- Open Rstudio and go to the unzipped folder
2. Getting started with online Docker image
- Launch an R studio interface in an R docker along with bioconductor packages for proteomics.